果胶是高等植物植物细胞壁的重要组成部分,它主要存在于初生壁和中间层中,次生壁及其他结构中也含有少量的果胶[1-2]。植物细胞壁中的果胶主要包括四种类型:同聚半乳糖醛酸(Homo- galacturonan,HG)、鼠李糖半乳糖醛酸I(Rhamno- galacturonan I,RG-I)、鼠李糖半乳糖醛酸II(Rhamno- galacturonan II,RG-II)以及木聚糖(Xylo gal- acturonan,XGA)等(图1A)[3-4],其中HG和RG-I分别约占总果胶含量的65%和20%~35%,RG-II和XGA含量均约为10%[2, 5]。
|
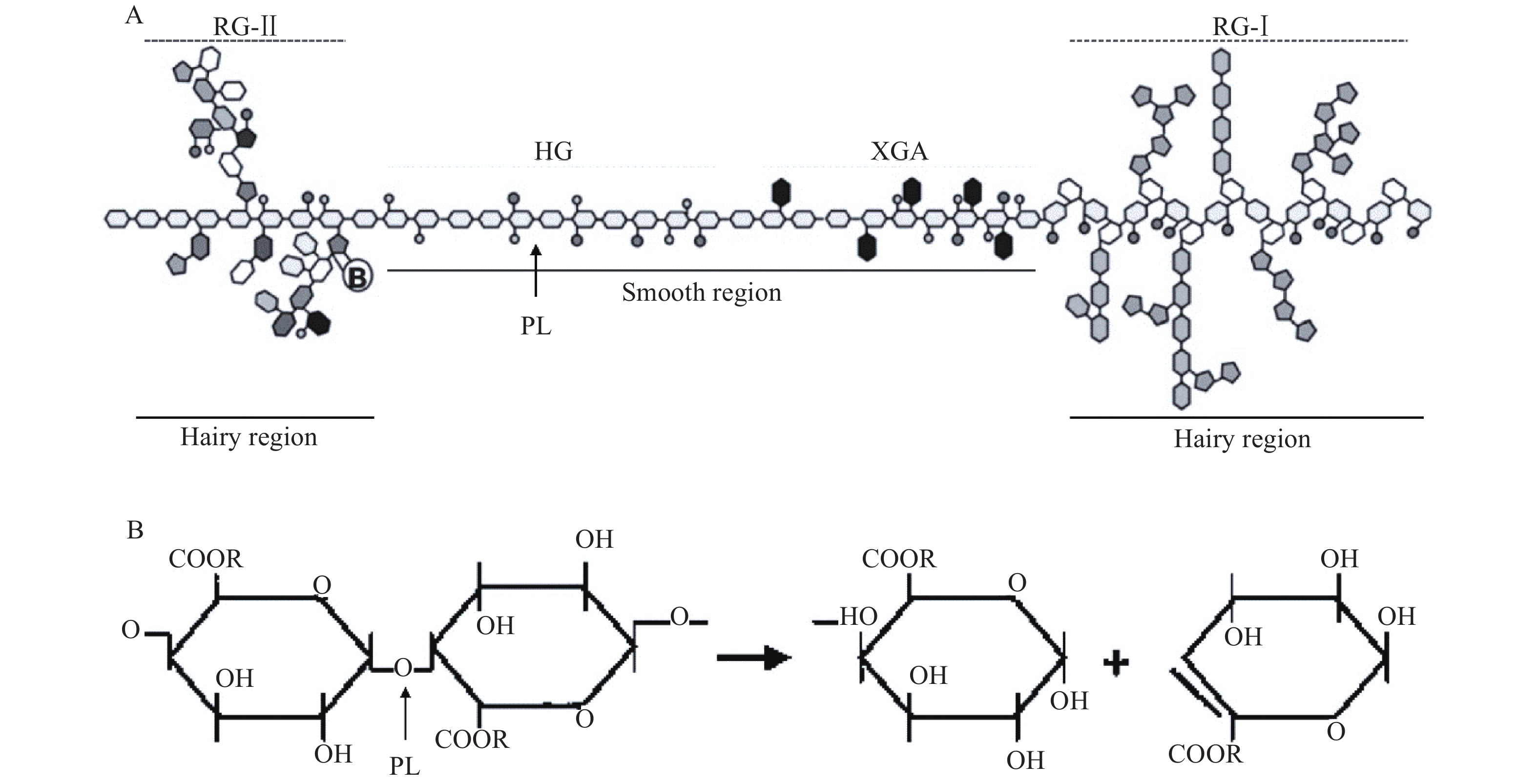
图 1 果胶结构示意图及果胶裂解酶作用方式[3-4] Fig. 1 Schematic structure of pectin and action mode of pectate lyase A:植物细胞壁中果胶结构示意图;B:果胶裂解酶降解去甲酯化果胶的作用方式 A: Schematic structure of pectin in plant cell wall; B: The action mode of pectate lyase on demethylated pectin |
果胶的代谢过程涉及多种酶类,其中果胶裂解酶(Pectate lyase,PL,EC 4.2.2.2)是一类能够通过β−反式消除作用催化降解位于果胶平滑区去甲酯化HG的α−1,4−糖苷键,在非还原性末端产生含有不饱和半乳糖醛酸残基的寡聚糖(图1B)[3, 6]。酶活性动力学测定显示,PL活性需要Ca2+、Mg2+或Mn2+等二价阳离子的参与,但对酚类化合物、硫醇、还原剂、碘酸盐和N−溴代琥珀酰亚胺的抑制作用比较敏感[7-10]。
PL的活性最早是在病原微生物中检测到的。研究显示,病原微生物通过分泌PL降解植物细胞壁以利于其入侵感染,但这一过程也会激发植物的免疫反应[11]。PL被广泛用于工业生产上,如葡萄酒和果汁的萃取与纯化、棉麻纤维的脱胶与精炼、以及果胶废水的处理等方面[4, 6]。高等植物中果胶裂解酶基因PL(又称Pectate lyase like,PLL或PEL)最先是在番茄花粉中克隆得到的,随后陆续在其他植物中也鉴定到PL基因[6, 12]。蛋白质多序列比对分析显示,植物PL蛋白均具有一个保守Pec_Lyase_C结构域,部分PL蛋白具有一个信号肽和一个位于N端的Pec_Lyase_N结构域[13-17]。
1 植物PL家族的分布及表达随着多种植物全基因组测序的完成,不同植物的PL基因家族成员也得到广泛的鉴定,且这些基因呈现出多样化的表达模式。双子叶植物水稻Oryza sativa基因组上预测有12个OsPLL基因;只有OsPLL1、OsPLL3、OsPLL4和OsPLL12这4个基因在茎、叶鞘和小穗中检测到有表达,且在花粉发育的各个时期均有表达[14]。
单子叶植物拟南芥Arabidopsis thaliana中有26个AtPLL基因,所有AtPLL基因均在花器官中表达且大多数基因在花器官中的表达量高于其他组织,AtPLL3、AtPLL7和AtPLL4只在花器官中表达但不在花粉中表达,10个AtPLL基因在根、茎、叶和花中均有表达;启动子表达分析显示,大多数的AtPLL基因主要在花器官和种子的离层区、果实的开裂区等细胞分离的局部区域表达,少部分在花柱和侧根发生时的内皮层、皮质层等促进分离的细胞类型中表达[15, 18]。芜菁Brassica rapa中有46个BrPLL基因,其中26个BrPLL基因在所有组织器官中均表达,41个BrPLL基因在花器官中表达且有11个BrPLL基因只在花器官中表达,3个BrPLL基因在茎、叶、花和角果中表达而在根部不表达,BrPLL12-1只在花和角果中表达[16]。杨树Populus trichocarpa基因组上存在30个PtPL1基因,大多数PtPL1基因在雌蕊、雄蕊、根、幼叶和木质部中高表达,6个PtPL1基因主要在根部和木质部中表达,6个PtPL1基因在茎部不同部位均呈现高水平表达,2个PtPL1基因主要在木质部中表达[17, 19]。番茄Solanum lycopersicum基因组存在22个SlPL基因,有7个SlPL基因在花器官中呈现高水平的表达[20]。3种不同棉花品种Gossypium raimondii、G. arboreum和G. hirsutum基因组上分别有53、42和83个PEL基因;有32个GhPEL基因主要在花器官尤其是雄蕊中表达,可能参与调控花器官的发育;在各个组织中均检测到表达的16个GhPEL基因中有7个主要在棉花纤维中表达,表明这些基因可能参与调控棉花纤维的发育;另外,有4个GhPEL基因主要在叶片中表达,13个GhPEL基因主要在营养生长和生殖生长的器官中表达,其表达量比棉纤维中的高;且经植物生长素(Indole-3-acetic acid,IAA)处理后,7个GhPEL基因明显上调表达,8个GhPEL基因的表达受到抑制,GhPEL67则呈现出先下调后上调的表达模式,表明生长素可能通过调控GhPEL基因参与生长发育途径[21]。
2 植物PL的生物学功能研究表明,植物PL具有多种生物学功能,主要参与植物的生长发育、花器官的发育、果实的成熟软化、与病原微生物的相互作用等不同的生理过程,同时也能够引起动植物的过敏性反应。
2.1 PL参与植物的生长发育植物细胞的伸长能力受到细胞壁的限制,因而,植物细胞壁中果胶的代谢会影响细胞的松弛和扩张,从而影响植株的发育[22]。
拟南芥AtPLA1和AtPLA2在侧根发生时受到IAA诱导上调表达,表明其可能参与侧根形成;另外AtPLL1、AtPLL12、AtPLL13、AtPLL23和AtPLL26受到生长素的诱导表达,可能参与细胞伸长和分化[14, 23]。日本柳树Salix gilgiana的SgPEL1在植株中呈现组成型表达,主要在幼嫩叶片和茎中、以及在雄蕊的延伸花丝、雌蕊的分泌组织、花絮的木质部薄壁组织中表达;生长素Auxin可以快速诱导百日草Zinnia elegans导管组织Zepel的表达,且Zepel的表达与维管束和腋芽原基相关;表明这些PL基因可能参与细胞伸长与分化过程中细胞壁的重塑[24-25]。
拟南芥PMR6突变后导致植物细胞壁成分发生改变,并使植株的营养生长受到抑制;AtPL18或AtPL19突变后会使植株根部和叶片的生长受到抑制[26-27]。水稻的OsPSE1是第1个被发现与植株衰老有关的基因。该基因突变后会导致植株进入生殖生长期后,快速进入衰老期,缩短了水稻生长期,影响水稻光合能量转化与积累,种子灌浆不充实从而导致水稻减产[28];最近克隆的另一个水稻DEL1基因是调控机制最清晰的PL基因。DEL1主要在植株伸长组织中表达,它突变后会降低PL活性,增加甲酯化HG含量,从而改变细胞壁成分和结构,影响植株的生长和衰老[29]。杨树PtPL1−18主要次生壁形成早期的叶柄的维管束、茎部的初生木质部和维管束间表达,PtxtPL1−27主要在次生壁形成后期分化的木质部中表达;PtPL1−18的过量表达会导致茎组织木质部果胶含量降低、纤维素和半纤维素以及可溶性糖含量增高,降低白杨次级细胞壁的厚度,影响木质部细胞的形态;而PtxtPL1−27的过量表达能够提高PL活性,促进木质纤维素糖化,增加单糖特别是木糖产量;从而参与杨树的生长及木质部的发育过程[17, 30]。棉花G. hirsutum的GhPEL主要在棉花开花后的纤维中表达,抑制GhPEL的表达会影响其降解棉纤维初生壁中甲酯化果胶的能力,阻止组织细胞壁的松弛而抑制棉纤维的发育与伸长[31]。
2.2 PL参与植物花器官的发育植物花器官发育过程中涉及细胞壁中果胶、纤维素和半纤维素等成分的代谢及重塑,研究表明PL也参与花器官的发育过程[32]。
番茄LAT56和LAT59在成熟花粉和花药中高度表达,烟草TP10在花粉发育过程中的小孢子期特异表达,另外玉米和苜蓿中的PL基因均在花粉中特异表达,暗示着其参与植物花粉的发育过程[12, 33-35]。研究表明,位于LAT56的5'UTR片段抑制其表达水平,且LAT56蛋白受到糖基化修饰并定位于花粉细胞壁[34, 36]。乙烯可以诱导激活玫瑰Rosa bourboniana花瓣2~3细胞层区域的分离区细胞分离过程,RbPel1在花瓣分离区的表达量受到乙烯的诱导表达,表明乙烯激活PL基因可能参与了玫瑰花瓣脱落过程中间层的溶解从而促进花瓣的脱落[37]。
水稻OsPLL3和OsPLL4除了在茎和叶鞘中表达,且在花粉发育过程中呈现较高水平的表达;OsPLL4的沉默表达会使二核晚期的小孢子降解而形态畸形,OsPLL3的沉默表达会使花粉开裂期的成熟花粉降解,从而导致花粉败育,影响植株的花器官和小穗发育[14]。小白菜Brassica campestris的BcPLL9和BcPLL10主要在成熟花粉的小孢子和授粉过程的雌蕊柱头中表达;抑制BcPLL9和BcPLL10的表达会扰乱果胶代谢,影响花粉管内外壁的形成,并阻碍花粉管的生长,从而导致植株育性下降、降低种子收获量[38-39]。
2.3 PL在植物果实成熟和软化中的功能果实的成熟软化是决定其保质期和商业价值的主要因素,诸多研究表明,植物PL能够通过降解细胞壁中的去甲酯化果胶参与果实的成熟软化过程[40]。
日本杏子Prunus mume的Pm65在果实成熟软化过程的果皮中上调表达;桃树Prunus persica 中PpPL1、PpPL2、PpPL3、PpPL4和PpPL的表达量随着果实成熟及花色苷降解过程逐步上升;从成熟后的番木瓜果实中克隆到PL基因,暗示着这些PL基因参与了杏子、桃子和木瓜的成熟软化以及桃子果实的花色苷的降解过程[41-44]。芒果Mangifera indica PL活性和可溶性果胶含量随着果实的成熟而提高,基因表达分析显示MiPel1在果实成熟过程中特异表达且受到乙烯的诱导显著上调表达,而甲基环丙烯(1-Methylcyclopropene)处理后会明显推迟其表达,暗示着MiPel1参与果实成熟过程中果胶的降解从而促进果实软化[7]。香蕉Musa acuminata的MaPel1、MaPel2、MWPL1、MWPL2在果实中特异表达,植物激素如2,4−二氯苯氧乙酸(2,4-Dichlorophenoxy acetic acid,2,4-D)、乙烯、赤霉素处理均可以诱导香蕉果实中的果胶裂解酶基因和果胶裂解酶活性[6, 24, 45-47]。
番茄SlPL、SlPL5、SlPL16和SlPL19在果实成熟过程中下调表达,但SlPL在果实软化过程中显著上调表达且受到1−氨基环丙烷−1−羧酸(1-Aminocyclopropane-1-carboxylic acid,ACC)、脱落酸(Abscisic acid,ABA)、IAA和乙烯等激素的诱导表达[19]。抑制SlPL的表达会导致果实中果胶含量降低,纤维素和半纤维素含量提高,并改变果皮细胞的大小、数目及厚度从而提高果实硬度;另外超氧化物歧化酶、过氧化物酶和过氧化氢酶活性的提高可以提高果实的抗氧化能力,从而抑制果实的软化并提高果实的储藏寿命[19, 48]。
森林草莓Fragaria vesca的FvPLA、FvPLB、FvPLC和智利草莓F. chiloensis的FcPL1以及凤梨草莓F. ×ananassa的FaplA、FaplB在果实成熟和软化过程中表达量逐渐上升,且果胶裂解酶活性随着果实硬度下降而上升,说明其可能参与草莓果实的软化过程[49-52]。抑制草莓PL基因的表达可以减少果实软化过程中细胞壁的初生壁和中间层中果胶的解聚程度,提高果实的硬度从而有效提高其储藏能力[53-57]。研究表明,高浓度CO2短暂处理后可以增加草莓果实硬度,且草莓中的PL活性受到抑制,说明CO2可能通过抑制草莓果实中PL活性防止其降解细胞壁中的果胶从而增加果实的硬度[49, 58]。
2.4 PL参与植物与病原微生物的相互作用细胞壁是植物抵御病原微生物侵染的第1道屏障,许多研究表明,病原微生物通过释放果胶裂解酶降解植物细胞壁以利于入侵[11, 59]。但近些年的一些研究表明,植物的果胶裂解酶同样参与了植物与病原微生物之间的相互作用。
拟南芥PMR6突变后改变植株细胞壁成分,提高植株对白粉病的抗性,说明PMR6受到病原菌的利用参与植物对病原微生物的感病反应[28]。灰霉菌Botrytis cinerea侵染会抑制番茄SlPL和SlPL5的表达,降低SlPL的表达可以提高果实对B. cinerea的抗性从而延缓果实的腐烂[17]。黄单孢杆菌Xanthomonas gardneri分泌的转录激活因子样效应物(Transcription activator-like effector, TALE)AvrHaH1蛋白会导致番茄果实和叶片产生水浸病害;研究表明AvrHaH1能够结合番茄bHLH(Basic helix-loop-helix)转录因子的效应蛋白结合元件(Effector binding elements,EBEs)诱导bHLH基因的表达,激活的bHLH进一步结合PL基因Solyc05g014000的启动子诱导其表达;而PL基因的表达可能通过降解植物细胞壁中果胶而增加组织浸软性[60]。
囊肿线虫Heterodera schachtii和根结线虫Meloidogyne incognita侵染拟南芥后,会在侵染部位分别诱导产生合胞体和大细胞为线虫寄生提供场所,拟南芥AtPL18和AtPL19在合胞体或大细胞中均被诱导上调表达;AtPL18和AtPL19分别突变后会使囊肿线虫诱导的合胞体中雌性囊肿线虫数量减少,但不影响根结线虫诱导的大细胞中线虫的数量;且在囊肿线虫侵染早期合胞体和合胞体细胞壁中甲酯化HG含量低于侵染后期,而侵染早期去甲酯化HG含量则高于侵染后期;表明AtPL18和AtPL19可能通过降解植物细胞壁中的甲酯化HG而参与囊肿线虫侵染早期合胞体的形成[27]。
根瘤菌Mesorhizobium loti与豆科植物相互作用形成根瘤,而根瘤菌会在侵染部位降解植物细胞壁并形成侵染线入侵根瘤[61]。百脉根Lotus japonicus结瘤上的LjNPL在根部和根毛的表达受到根瘤菌结瘤因子(Nod)的诱导表达,LjNPL突变后影响成熟感染根瘤的形成,参与根瘤形成和侵染过程的NIN转录因子能够结合到LjNPL的启动子上;表明根瘤菌通过激活结瘤信号通路和NIN转录因子诱导LjNPL的表达,以降解植物细胞壁从而形成成熟的根瘤[62]。大豆Glycine max囊泡相关膜蛋白GmVAMP721d突变后影响根瘤菌感染细胞中甲酯化和去甲酯化果胶的分布,并且GmVAMP721d与果胶裂解酶GmNPL1共定位于侵染线的囊泡中,表明GmVAMP721d可能通过参与GmNPL1的转运从而参与侵染线中根瘤菌的释放[63]。
2.5 PL引起动植物的过敏性反应来源于菊科Asteraceae、柏科Cupressaceae等多种植物的过敏性成员释放的花粉会诱发哺乳动物呼吸道的过敏性反应[64-65]。
菊科植物豚草Ambrosia artemisiifolia的Amb a 1、艾草Artemisia vulgaris的Art v 6,以及柏科植物日本扁柏Chamaecyparis obtusa的Cha o 1、日本柳杉Cryptomeria japonica的Cry j I、绿干柏Cupressus arizonica的Cup a 1、地中海柏木Cupressus sempervirens的Cup s 1、美国雪松Juniperus ashei的Jun a 1、大果刺柏Juniperus oxicedrus的Jun o 1、北美圆柏Juniperus virginiana的Jun v 1等引起哺乳动物过敏性反应的过敏原均属于PL家族成员[64, 66]。研究发现,菊科和柏科之间的PL过敏原不能引起交叉感染,但菊科内部或柏科内部的PL过敏原能引起交叉感染,为临床治疗由花粉引起过敏性反应提供重要参考依据[64]。另外,酶活性试验证实Cry j I、Jun a 1具有果胶裂解酶活性,通过烟草花叶病毒载体介导将Jun a 1导入烟草中,发现Jun a 1同样能够引起烟草的坏死症状[67-68]。
3 展望人们对植物PL基因的研究已经有30多年。早期的研究主要集中在PL基因的分离鉴定及表达分析上,仅有少数研究发现通过基因沉默技术将PL敲减能有效提高果实的硬度。随着生子生物学技术的快速发展以及大规模植物基因组测序的完成,关于植物PL家族的鉴定及功能将得到更为深入的研究。研究显示,PL基因广泛存在于植物基因组中,表达模式呈现多样化,参与植物生长发育、花器官发育、果实的成熟软化、植物与病原菌的相互作用、以及对动物产生过敏性反应等不同的生理过程。但植物PL基因通过何种分子机制和调控网络发挥生物学功能的研究仍然面临着巨大的挑战,当中最大的困难在于植物基因组存在大量的PL成员。这些PL功能相对保守,很可能存在功能冗余,导致单个基因突变的表型不明显。当前,出现的多基因编辑系统,有可能实现同时对植物多个PL基因进行敲除,从而推动该基因家族分子机制的研究。
| [1] |
MOHNEN D. Pectin structure and biosynthesis[J]. Curr Opin Plant Biol, 2008, 11(3): 266-277. DOI:10.1016/j.pbi.2008.03.006 (  0) 0) |
| [2] |
HARHOLT J, SUTTANGKAKUL A, VIBE S H. Biosynthesis of pectin[J]. Plant Physiol, 2010, 153(2): 384-395. DOI:10.1104/pp.110.156588 (  0) 0) |
| [3] |
YADAV S, YADAV P K, YADAV D, et al. Pectin lyase: A review[J]. Process Biochem, 2009, 44(1): 1-10. DOI:10.1016/j.procbio.2008.09.012 (  0) 0) |
| [4] |
JAYANI R S, SAXENA S, GUPTA R. Microbial pectinolytic enzymes: A review[J]. Process Biochem, 2005, 40(9): 2931-2944. DOI:10.1016/j.procbio.2005.03.026 (  0) 0) |
| [5] |
ATMODJO M A, HAO Z, MOHNEN D. Evolving views of pectin biosynthesis[J]. Annu Rev Plant Biol, 2013, 64: 747-779. DOI:10.1146/annurev-arplant-042811-105534 (  0) 0) |
| [6] |
MARIN-RODRIGUEZ M C, ORCHARD J, SEYMOUR G B. Pectate lyases, cell wall degradation and fruit softening[J]. J Exp Bot, 2002, 53(377): 2115-2119. DOI:10.1093/jxb/erf089 (  0) 0) |
| [7] |
MARIN-RODRIGUEZ M C, SMITH D L, MANNING K, et al. Pectate lyase gene expression and enzyme activity in ripening banana fruit[J]. Plant Mol Biol, 2003, 51(6): 851-857. DOI:10.1023/A:1023057202847 (  0) 0) |
| [8] |
CHOURASIA A, SANE V A, NATH P. Differential expression of pectate lyase during ethylene-induced postharvest softening of mango (Mangifera indica var. Dashehari)
[J]. Physiol Plantarum, 2006, 128(3): 546-555. (  0) 0) |
| [9] |
PAYASI A, MISRA P C, SANWAL G G. Purification and characterization of pectate lyase from banana (Musa acuminata) fruits
[J]. Phytochemistry, 2006, 67(9): 861-869. DOI:10.1016/j.phytochem.2006.02.003 (  0) 0) |
| [10] |
HERRON S R, SCAVETTA R D, GARRETT M, et al. Characterization and implications of Ca2+ binding to pectate lyase C
[J]. J Biol Chem, 2003, 278(14): 12271-12277. DOI:10.1074/jbc.M209306200 (  0) 0) |
| [11] |
LIONETTI V, CERVONE F, BELLINCAMPI D. Methyl esterification of pectin plays a role during plant-pathogen interactions and affects plant resistance to diseases[J]. J Plant Physiol, 2012, 169(16): 1623-1630. DOI:10.1016/j.jplph.2012.05.006 (  0) 0) |
| [12] |
WING R A, YAMAGUCHI J, LARABELL S K, et al. Molecular and genetic characterization of two pollen-expressed genes that have sequence similarity to pectate lyases of the plant pathogen Erwinia
[J]. Plant Mol Biol, 1990, 14(1): 17-28. DOI:10.1007/BF00015651 (  0) 0) |
| [13] |
DUBEY A K, YADAV S, KUMAR M, et al. In silico characterization of pectate lyase protein sequences from different source organisms[J]. Enzyme Res, 2010, 2010: 950230. (  0) 0) |
| [14] |
ZHENG Y, YAN J, WANG S, et al. Genome-wide identification of the pectate lyase-like (PLL) gene family and functional analysis of two PLL genes in rice
[J]. Mol Genet Genomics, 2018, 293(6): 1317-1331. DOI:10.1007/s00438-018-1466-x (  0) 0) |
| [15] |
SUN L, VAN NOCKER S. Analysis of promoter activity of members of the PECTATE LYASE-LIKE (PLL) gene family in cell separation in Arabidopsis
[J]. BMC Plant Biol, 2010, 10: 152. DOI:10.1186/1471-2229-10-152 (  0) 0) |
| [16] |
JIANG J, YAO L, MIAO Y, et al. Genome-wide characterization of the pectate lyase-like (PLL) genes in Brassica rapa
[J]. Mol Genet Genomics, 2013, 288(11): 601-614. DOI:10.1007/s00438-013-0775-3 (  0) 0) |
| [17] |
BAI Y, WU D, LIU F, et al. Characterization and functional analysis of the poplar pectate lyase-like gene PtPL1-18 reveal its role in the development of vascular tissues
[J]. Front Plant Sci, 2017, 8: 1123. DOI:10.3389/fpls.2017.01123 (  0) 0) |
| [18] |
PALUSA S G, GOLOVKIN M, SHIN S B, et al. Organ-specific, developmental, hormonal and stress regulation of expression of putative pectate lyase genes in Arabidopsis
[J]. New Phytol, 2007, 174(3): 537-550. DOI:10.1111/nph.2007.174.issue-3 (  0) 0) |
| [19] |
GEISLER-LEE J, GEISLER M, COUTINHO P M, et al. Poplar carbohydrate-active enzymes. Gene identification and expression analyses[J]. Plant Physiol, 2006, 140(3): 946-962. DOI:10.1104/pp.105.072652 (  0) 0) |
| [20] |
YANG L, HUANG W, XIONG F, et al. Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould
[J]. Plant Biotechnol J, 2017, 15(12): 1544-1555. DOI:10.1111/pbi.2017.15.issue-12 (  0) 0) |
| [21] |
SUN H, HAO P, MA Q, et al. Genome-wide identification and expression analyses of the pectate lyase (PEL) gene family in cotton (Gossypium hirsutum L.)
[J]. BMC Genomics, 2018, 19(1): 661. DOI:10.1186/s12864-018-5047-5 (  0) 0) |
| [22] |
WOLF S, MOUILLE G, PELLOUX J. Homogalacturonan methyl-esterification and plant development[J]. Mol Plant J, 2009, 2(5): 851-860. DOI:10.1093/mp/ssp066 (  0) 0) |
| [23] |
LASKOWSKI M, BILLER S, STANLEY K, et al. Expression profiling of auxin-treated Arabidopsis roots: Toward a molecular analysis of lateral root emergence
[J]. Plant Cell Physiol, 2006, 47(6): 788-792. DOI:10.1093/pcp/pcj043 (  0) 0) |
| [24] |
PUA E, ONG C, LIU P, et al. Isolation and expression of two pectate lyase genes during fruit ripening of banana (Musa acuminata)
[J]. Physiologia Plantarum, 2001, 113(1): 92-99. DOI:10.1034/j.1399-3054.2001.1130113.x (  0) 0) |
| [25] |
DOMINGO C, ROBERTS K, STACEY N J, et al. A pectate lyase from Zinnia elegans is auxin inducible
[J]. Plant, 1998, 13(1): 17-28. (  0) 0) |
| [26] |
WIECZOREK K, ELASHRY A, Quentin M, et al. A distinct role of pectate lyases in the formation of feeding structures induced by cyst and root-knot nematodes[J]. Mol Plant Microbe Interact, 2014, 27(9): 901-912. DOI:10.1094/MPMI-01-14-0005-R (  0) 0) |
| [27] |
VOGEL J P, RAAB T K, SCHIFF C, et al. PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis
[J]. Plant Cell, 2002, 14(9): 2095-2106. DOI:10.1105/tpc.003509 (  0) 0) |
| [28] |
LENG Y, YANG Y, REN D, et al. A rice PECTATE LYASE-LIKE gene is required for plant growth and leaf senescence[J]. Plant Physiol, 2017, 174(2): 1151-1166. DOI:10.1104/pp.16.01625 (  0) 0) |
| [29] |
WU H B, WANG B, CHEN Y, et al. Characterization and fine mapping of the rice premature senescence mutant ospse1
[J]. Theor Appl Genet, 2013, 126(7): 1897-1907. DOI:10.1007/s00122-013-2104-y (  0) 0) |
| [30] |
BISWAL A K, SOENO K, GANDLA M L, et al. Aspen pectate lyase PtxtPL1-27 mobilizes matrix polysaccharides from woody tissues and improves saccharification yield[J]. Biotechnol Biofuels, 2014, 7(1): 11. DOI:10.1186/1754-6834-7-11 (  0) 0) |
| [31] |
WANG H, GUO Y, LÜ F, et al. The essential role of GhPEL gene, encoding a pectate lyase, in cell wall loosening by depolymerization of the de-esterified pectin during fiber elongation in cotton
[J]. Plant Mol Biol, 2010, 72(4/5): 397-406. (  0) 0) |
| [32] |
MOLLET J C, LEROUX C, DARDELLE F, et al. Cell wall composition, biosynthesis and remodeling during pollen tube growth[J]. Plants, 2013, 2(1): 107-147. DOI:10.3390/plants2010107 (  0) 0) |
| [33] |
TURCICH M P, HAMILTON D A, MASCARENHAS J P. Isolation and characterization of pollen-specific maize genes with sequence homology to ragweed allergens and pectate lyases[J]. Plant Mol Biol, 1993, 23(5): 1061-1065. DOI:10.1007/BF00021820 (  0) 0) |
| [34] |
DIRCKS L K, VANCANNEYT G, MCCORMICK S. Biochemical characterization and baculovirus expression of the pectate lyase-like LAT56 and LAT59 pollen proteins of tomato[J]. Plant Physiol Bioch, 1996, 34(4): 509-520. (  0) 0) |
| [35] |
WU Y, QIU X, DU S, et al. PO149, a new member of pollen pectate lyase-like gene family from alfalfa
[J]. Plant Mol Biol, 1996, 32(6): 1037-1042. DOI:10.1007/BF00041387 (  0) 0) |
| [36] |
CURIE C, MCCORMICK S. A strong inhibitor of gene expression in the 5' untranslated region of the pollen-specific LAT59 gene of tomato
[J]. Plant Cell, 1997, 9(11): 2025-2036. (  0) 0) |
| [37] |
SINGH A P, PANDEY S P, RAJLUXMI, et al. Transcriptional activation of a pectate lyase gene, RbPel1, during petal abscission in rose
[J]. Postharvest Biol Tec, 2011, 60(2): 143-148. DOI:10.1016/j.postharvbio.2010.12.014 (  0) 0) |
| [38] |
JIANG J, YAO L, YU Y, et al. PECTATE LYASE-LIKE10 is associated with pollen wall development in Brassica campestris
[J]. J Integr Plant Biol, 2014, 56(11): 1095-1105. DOI:10.1111/jipb.v56.11 (  0) 0) |
| [39] |
JIANG J, YAO L, YU Y, et al. PECTATE LYASE-LIKE 9 from Brassica campestris is associated with intine formation
[J]. Plant Sci, 2014, 229: 66-75. DOI:10.1016/j.plantsci.2014.08.008 (  0) 0) |
| [40] |
WANG D, YEATS T H, ULUISIK S, et al. Fruit softening: Revisiting the role of pectin[J]. Trends Plant Sci, 2018, 23(4): 302-310. DOI:10.1016/j.tplants.2018.01.006 (  0) 0) |
| [41] |
MITA S, NAGAI Y, ASAI T. Isolation of cDNA clones corresponding to genes differentially expressed in pericarp of mume (Prunus mume) in response to ripening, ethylene and wounding signals
[J]. Physiologia Plantarum, 2006, 128(3): 531-545. (  0) 0) |
| [42] |
林莹, 陈晓静. 番木瓜果胶裂解酶基因片段的克隆及序列分析[J]. 漳州师范学院学报, 2009, 22(1): 84-87. DOI:10.3969/j.issn.1008-7826.2009.01.021 (  0) 0) |
| [43] |
袁成龙. 桃果实果胶裂解酶基因PpPLs的克隆及表达分析[D]. 青岛: 青岛农业大学, 2013.
(  0) 0) |
| [44] |
王柳凤, 樊连梅, 刘更森, 等. 乌桃果胶裂解酶基因(PpPL)的克隆及其在花色苷降解中的作用
[J]. 分子植物育种, 2017, 15(8): 3046-3056. (  0) 0) |
| [45] |
DOMINGUEZ-PUIGJANER E, LLOP I, VENDRELL M, et al. A cDNA clone highly expressed in ripe banana fruit shows homology to pectate lyases[J]. Plant Physiol, 1997, 114(3): 1071-1076. DOI:10.1104/pp.114.3.1071 (  0) 0) |
| [46] |
PAYASI A, MISRA P C, SANWAL G G. Effect of phytohormones on pectate lyase activity in ripening Musa acuminata
[J]. Plant Physiol Bioch, 2004, 42(11): 861-865. DOI:10.1016/j.plaphy.2004.10.011 (  0) 0) |
| [47] |
PAYASI A, SANWAL G G. Pectate lyase activity during ripening of banana fruit[J]. Phytochemistry, 2003, 63(3): 243-248. DOI:10.1016/S0031-9422(03)00027-X (  0) 0) |
| [48] |
ULUISIK S, CHAPMAN N H, SMITH R, et al. Genetic improvement of tomato by targeted control of fruit softening[J]. Nat Biotechnol, 2016, 34(9): 950-952. DOI:10.1038/nbt.3602 (  0) 0) |
| [49] |
BENITEZ-BURRACO A, BLANCO-PORTALES R, REDONDO-NEVADO J, et al. Cloning and characterization of two ripening-related strawberry (Fragaria x ananassa cv. Chandler) pectate lyase genes
[J]. J Exp Bot, 2003, 54(383): 633-645. DOI:10.1093/jxb/erg065 (  0) 0) |
| [50] |
FIGUEROA C R, PIMENTEL P, GAETE-EASTMAN C, et al. Softening rate of the Chilean strawberry (Fragaria chiloensis) fruit reflects the expression of polygalacturonase and pectate lyase genes
[J]. Postharvest Biol Tec, 2008, 49(2): 210-220. DOI:10.1016/j.postharvbio.2008.01.018 (  0) 0) |
| [51] |
周鹤莹, 张玮, 张卿, 等. 森林草莓‘Ruegen’果胶裂解酶基因的克隆及荧光定量表达分析[J]. 园艺学报, 2015, 42(3): 455-461. (  0) 0) |
| [52] |
MEDINA-ESCOBAR N, CARDENAS J, MOYANO E, et al. Cloning, molecular characterization and expression pattern of a strawberry ripening-specific cDNA with sequence homology to pectate lyase from higher plants[J]. Plant Mol Biol, 1997, 34(6): 867-877. DOI:10.1023/A:1005847326319 (  0) 0) |
| [53] |
SESMERO R, QUESADA M A, MERCADO J A. Antisense inhibition of pectate lyase gene expression in strawberry fruit: Characteristics of fruits processed into jam[J]. J Food Eng, 2007, 79(1): 194-199. DOI:10.1016/j.jfoodeng.2006.01.044 (  0) 0) |
| [54] |
POSE S, KIRBY A R, PANIAGUA C, et al. The nanostructural characterization of strawberry pectins in pectate lyase or polygalacturonase silenced fruits elucidates their role in softening[J]. Carbohydr Polym, 2015, 132: 134-145. DOI:10.1016/j.carbpol.2015.06.018 (  0) 0) |
| [55] |
SANTIAGO-DOMENECH N, JIMENEZ-BEMUDEZ S, MATAS A J, et al. Antisense inhibition of a pectate lyase gene supports a role for pectin depolymerization in strawberry fruit softening[J]. J Exp Bot, 2008, 59(10): 2769-2779. DOI:10.1093/jxb/ern142 (  0) 0) |
| [56] |
YOUSSEF S M, JIMENEZ-BERMUDEZ S, BELLIDO M L, et al. Fruit yield and quality of strawberry plants transformed with a fruit specific strawberry pectate lyase gene[J]. Sci Hortic-Amsterdam, 2009, 119(2): 120-125. DOI:10.1016/j.scienta.2008.07.011 (  0) 0) |
| [57] |
JIMENEZ-BERMUDEZ S, REDONDO-NEVADO J, MUNOZ-BLANCO J, et al. Manipulation of strawberry fruit softening by antisense expression of a pectate lyase gene[J]. Plant Physiol, 2002, 128(2): 751-759. DOI:10.1104/pp.010671 (  0) 0) |
| [58] |
WANG M H, KIM J G, AHN S E, et al. Potential role of pectate lyase and Ca2+ in the increase in strawberry fruit firmness induced by short-term treatment with high-pressure CO2[J]. J Food Sci, 2014, 79(4): s685-s692. DOI:10.1111/jfds.2014.79.issue-4 (  0) 0) |
| [59] |
HEMATY K, CHERK C, SOMERVILLE S. Host-pathogen warfare at the plant cell wall[J]. Curr Opin Plant Biol, 2009, 12(4): 406-413. DOI:10.1016/j.pbi.2009.06.007 (  0) 0) |
| [60] |
SCHWARTZ A R, MORBITZER R, LAHAYE T, et al. TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato[J]. Proc Natl Acad Sci USA, 2017, 114(5): e897-e903. DOI:10.1073/pnas.1620407114 (  0) 0) |
| [61] |
KONDOROSI E, MERGAERT P, KERESZT A. A paradigm for endosymbiotic life: Cell differentiation of Rhizobium bacteria provoked by host plant factors[J]. Annu Rev Microbiol, 2013, 67: 611-628. DOI:10.1146/annurev-micro-092412-155630 (  0) 0) |
| [62] |
XIE F, MURRAY J D, KIM J, et al. Legume pectate lyase required for root infection by rhizobia[J]. Proc Natl Acad Sci USA, 2012, 109(2): 633-638. DOI:10.1073/pnas.1113992109 (  0) 0) |
| [63] |
GAVRIN A, CHIASSON D, OVCHINNIKOVA E, et al. VAMP721a and VAMP721d are important for pectin dynamics and release of bacteria in soybean nodules[J]. New Phytol, 2016, 210(3): 1011-1021. DOI:10.1111/nph.13837 (  0) 0) |
| [64] |
PICHLER U, HAUSER M, WOLF M, et al. Pectate lyase pollen allergens: Sensitization profiles and cross-reactivity pattern[J]. PLoS One, 2015, 10(5): e120038. (  0) 0) |
| [65] |
GADERMAIER G, HAUSER M, FERREIRA F. Allergens of weed pollen: An overview on recombinant and natural molecules[J]. Methods, 2014, 66(1): 55-66. DOI:10.1016/j.ymeth.2013.06.014 (  0) 0) |
| [66] |
CHARPIN D, CALLEJA M, LAHOZ C, et al. Allergy to cypress pollen[J]. Allergy, 2005, 60(3): 293-301. DOI:10.1111/all.2005.60.issue-3 (  0) 0) |
| [67] |
LIU Z, BHATTACHARYYA S, NING B, et al. Plant-expressed recombinant mountain cedar allergen Jun a 1 is allergenic and has limited pectate lyase activity[J]. Int Arch Allergy Immunol, 2010, 153(4): 347-358. DOI:10.1159/000316345 (  0) 0) |
| [68] |
TANIGUCHI Y, ONO A, SAWATANI M, et al. Cry j I, a major allergen of Japanese cedar pollen, has pectate lyase enzyme activity[J]. Allergy, 1995, 50(1): 90-93. DOI:10.1111/all.1995.50.issue-1 (  0) 0) |
 2019, Vol. 40
2019, Vol. 40

