鸡肉是我国第二大肉类消费品,据世界粮农组织预测,至2021年,鸡肉有望成为世界第一大肉类供应来源[1]。骨骼肌是脊椎动物体内最大的组织,占整个机体总质量的40%左右,占全部蛋白质质量的50%~75%[2]。在家禽中,骨骼肌的生长发育与家禽的产肉量有着密切的联系,足量的骨骼肌是决定家禽肌肉产量的关键因素,因此骨骼肌生长发育过程备受瞩目。
骨骼肌作为躯体最重要的组成部分,其生长发育表现为肌细胞数量增加和体积扩大[3]。骨骼肌的生长发育涉及一系列复杂的生物学过程,主要包括间充质干细胞终末分化成为成肌细胞,成肌细胞增殖分化形成初级肌管,肌管成熟为肌纤维,肌纤维肥大4个阶段[4]。整个发育过程由多种要素共同参与,遗传因素发挥关键作用,受许多基因、信号通路和转录因子调控[5],如生肌调节因子(Myogenic regulatory factors,MRFs)[6]和肌细胞增强因子(Myocyte enhancer factor 2,MEF2)[7]等,肌肉特异性转录因子在很大程度上影响骨骼肌的正常生长发育。众多基因参与骨骼肌的生长发育,彼此互作构成复杂的调控网络维持骨骼肌生长发育稳定[8-9]。随着表观遗传学的发展,研究人员发现除了一些主效基因和转录因子之外,DNA甲基化、组蛋白修饰、非编码RNAs(Noncoding RNAs,ncRNAs)在骨骼肌生长发育过程中也发挥着重要的调控作用[10-12]。
ncRNAs是一类不编码蛋白质或者多肽,功能多样化,能够在RNA水平直接发挥调节作用的核苷酸,约85%的基因组可以转录成为ncRNAs。以长度、功能以及结构特征等作为划分依据,ncRNAs可分为小非编码RNAs(Small noncoding RNAs,small ncRNAs)如微小RNAs(Micro RNAs,miRNAs)、长链非编码RNAs(Long noncoding RNAs,lncRNAs)、环状RNAs(Circular RNAs,circRNAs)[13]。ncRNAs作为机体必需的调控因子,广泛参与各种生命活动,在细胞增殖、分化,重大疾病的发生及胚胎发育等过程中都具有关键性作用[14-17]。越来越多研究证明ncRNAs与骨骼肌发育过程(如成肌细胞增殖、分化和凋亡,肌纤维形成)及骨骼肌疾病密切相关[14-15, 18]。然而,目前仅有少数ncRNAs对骨骼肌的调节机制被研究清楚。近年来,关于miRNAs、lncRNAs及circRNAs在家禽骨骼肌生长发育过程中作用的研究越来越多。本文以此为基础,针对ncRNAs对家禽骨骼肌生长发育调控机制进行概述,为系统了解家禽生长发育的分子遗传机制提供理论参考。
1 miRNAs对家禽骨骼肌生长发育遗传调控的研究进展 1.1 miRNAs的特征与功能miRNAs是一类长度约22 nt,在转录后水平调控基因表达的内源性小非编码RNAs。miRNAs在生物体内广泛存在,可通过与靶基因的3′UTR完全或不完全互补配对,引起mRNA降解或翻译受阻,从而参与基因的表达调控[19],也可以结合靶基因的5′UTR、CDS区,从而在转录后水平调控基因表达[20-22]。miRNAs的表达具有组织特异性和时空特异性,在同一组织不同阶段、同一阶段不同组织的表达量及表达种类有较大区别。一种miRNA可以靶向多种mRNAs,一种mRNA也可以由多种miRNAs调控,这样miRNAs在生物体内构成一个极其精细的网络调控系统从而发挥重要作用[23]。研究发现miRNAs对细胞增殖、分化、融合和凋亡等基础过程具有一定的调控作用,在家禽体内,miRNAs主要与脂肪代谢[24]、肌肉发育[2]、胚胎发育[25]和疾病发生[17]等过程密切相关。调控骨骼肌发育的miRNAs可大致分为2类:一类是仅在肌肉组织特异性表达的miRNAs,被称作MyomiRs,另一类是在肌肉和其他组织细胞均可表达的miRNAs。试验表明不仅MyomiRs(如miR-206、miR-133和miR-1等)在家禽肌肉中特异性高表达,非MyomiRs在家禽骨骼肌发育过程中也具有调控功能[26-27]。
1.2 miRNAs的研究进展生长轴基因及其本身的遗传变异与家禽生长发育息息相关,针对这些基因的作用通路,学者们提出了多种假说。近年来,随着表观遗传学的发展,围绕“miRNA通过调控生长有关基因作用通路,进而影响家禽肌肉生长发育”展开了广泛研究。
早在2012年,利用表达谱芯片在14胚龄和7周龄正常鸡和矮小鸡骨骼肌中鉴定出差异表达的miRNA let-7b,let-7b不仅能靶向GHR基因调节骨骼肌的生长,也可以通过let-7b-IGF2BP3-IGF2通路抑制骨骼肌的生长发育,进一步研究证明该miRNA与矮小鸡的生长受阻有关[28]。Wang等[27, 29]在鸡肌肉中筛选到特异性表达的miR-133a,并发现miR-133a可能通过调控BIRC5的表达影响鸡骨骼肌的发育。2015年,Ouyang等[30]对快速生长型肉鸡(隐性白羽洛克)和慢速生长型肉鸡(杏花鸡)胸肌两尾样进行miRNA测序,发现了22个高表达丰度的差异表达miRNAs,靶基因预测及网络分析发现miR-34c、miR-223、miR-146b-3p、miR-21和miR-205a可能通过靶向生长相关基因影响鸡的生长。
近年来,研究发现了更多对成肌细胞的增殖、分化具有调控作用的miRNAs。miR-203是一个皮肤特异性miRNA,通过深度测序,Luo等[31]发现miR-203在鸡胚骨骼肌和鸡成肌细胞中均有表达,在10至16胚龄的骨骼肌中表达瞬时上调,而在16胚龄以后及成肌细胞分化成肌管期间表达明显下调,揭示该miRNA与肌肉发育相关,体外试验证明miR-203可通过靶向抑制c-JUN和MEF2C基因的表达抑制成肌细胞的增殖和分化。Wang等[32]结合转录组数据分析选取miR-205a和CDH11基因作为研究对象探讨二者在成肌细胞中发挥的作用,挖掘miR-205a自身表达调控的分子机制,发现miR-205a能够靶向抑制CDH11基因的表达,从而促进原代成肌细胞的增殖,抑制它们的分化。前人研究表明miR-206家族为肌肉特异性miRNA,在心肌和骨骼肌中高度富集,对肌肉细胞的增殖、分化、收缩和应激起到重要的调控作用[33],在家禽中研究发现启动子区域的SNP突变可直接影响鸡胚胎期肌肉生长[34]。2017年,Li等[35]在胚胎期骨骼肌组织中鉴定出高表达的miR-223,体外试验发现其可以靶向抑制IGF-2基因的表达,进而影响生长轴通路下游基因,调控肌肉细胞的增殖,MyoD也可以通过上调miR-223的表达来促进成肌细胞分化。而gga-miR-133a-3p和miR-34b-5p可以分别靶向PRRX1和IGFBP2基因调控成肌细胞的增殖和分化[36-37]。在肌肉发育过程中,E2F1的失调可抑制肌肉分化[38],在家禽中,E2F1能够与miR-20a-5p及miR-20b-5p相互联系形成负反馈环,实现对成肌细胞增殖与分化过程的调控[39]。
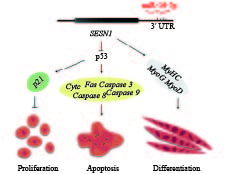
除了调节成肌细胞的增殖、分化外,miRNAs还可以通过它们的靶基因影响成肌细胞的凋亡等过程。2017年,在家禽中对miR-16家族研究发现,miR-16可以靶标抑制转录因子Foxo1和基因Bcl2,结合生长轴重要靶器官肌肉组织中高表达的TGFBR2等基因,调控肌肉细胞的增殖和凋亡[40]。miR-15a-16上游区域一段54 bp插入缺失可直接影响miR-16的生成,从而影响家鸡的肌肉量和体质量[41]。SESN1是应急诱导蛋白家族的一员,在肌肉损伤修复中起重要作用,它可以抑制肌肉萎缩,维持肌肉的完整性[42]。研究表明SESN1可以通过影响受损DNA修复,从而参与p53信号通路,最终调节成肌细胞的增殖与凋亡功能[43]。通过筛查快速生长型肉鸡和慢速生长型肉鸡miRNA表达谱发现miR-16-5p在两者之间差异表达,进一步研究发现miR-16-5p靶向抑制SESN1基因,调控p53信号通路,从而调控家鸡成肌细胞的增殖、分化和凋亡(图1)[44]。
|
图 1 miR-16-5p调控成肌细胞的增殖、分化和凋亡过程的作用机制 Fig. 1 Model of miR-16-5p regulating myoblast proliferation, apoptosis and differentiation |
lncRNAs指由RNA聚合酶Ⅱ合成,长度大于200 nt的非编码RNAs[45]。2002年,在小鼠中首次大规模发现lncRNAs[46]。lncRNAs数目繁多,功能复杂。近年来,越来越多的研究表明部分lncRNAs不仅在RNA水平起调节作用,还可以编码产生微肽,大大提高基因组的编码潜力,具有重要的进化意义[47]。lncRNAs可通过编码微肽,作为印记基因、分子海绵和调控转录因子活性等机制广泛参与肌肉生长发育的调控[14, 47-50]。
2.2 lncRNAs的研究进展linc-MD1是首个被鉴定的肌肉特异性lncRNA。2012年,研究人员运用转录组测序方法在家禽骨骼肌中鉴定发现了281个新lncRNAs[51]。随着测序技术的不断更新换代,数以千计与骨骼肌生长发育相关的lncRNAs被识别。近年来,基于多组学分析方法对与鸡肌肉生长发育相关的lncRNA研究发现lncRNA广泛参与家禽骨骼肌的调控网络。Li等[52]对杏花鸡11胚龄(E11)、16胚龄(E16)和孵化后1 d(D1)腿肌进行全转录组测序分析获得lncRNA和mRNA数据,发现了1 995个与骨骼肌有关的lncRNAs,其中部分lncRNAs可以通过顺式作用对靶标进行调控从而影响细胞生长增殖,例如,预测lnc00003323可能通过顺式作用机理靶向调控TEAD4表达。Ouyang等[53]联合分析E11、E16和D1 3个时期腿肌蛋白质组学数据和lncRNAs数据,发现4个与肌肉收缩过程有关的差异蛋白可能受lnc00068445、lnc00037615和lnc00037619的靶调控,影响胚胎期骨骼肌的生长发育。
由于lncRNA可通过不同的作用机制调节机体功能,在前期鉴定出的众多可能与骨骼肌生长发育有关的lncRNAs的基础上,研究人员从分子细胞水平对lncRNA调控家鸡肌肉生长发育的作用机制进行了深入探讨发现,lncRNA可以通过吸附miRNA或者编码短分子微肽调控重要功能基因的表达,从而调控基因作用通路来影响鸡肌肉的生长发育。
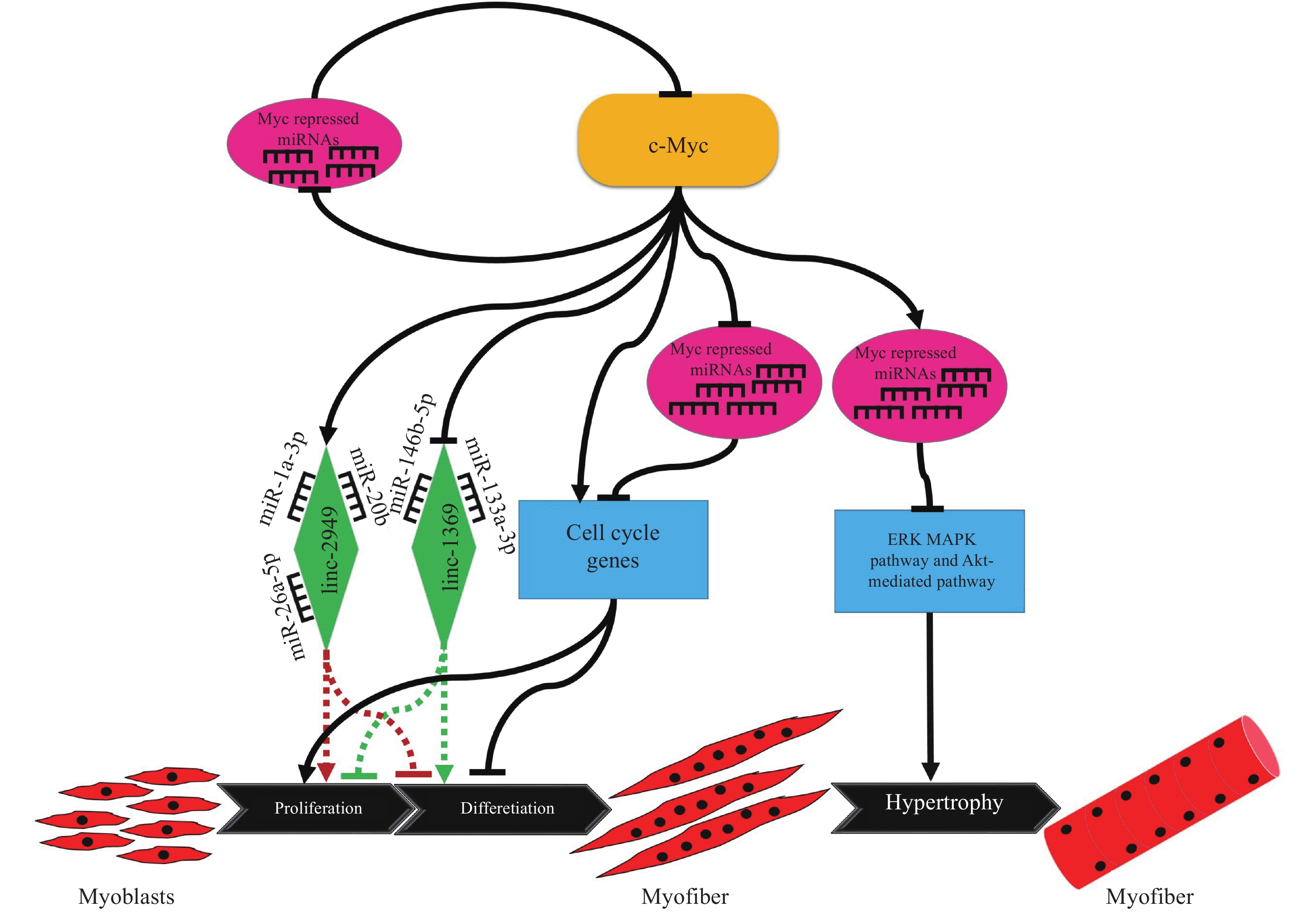
首先lncRNAs可以作为竞争性内源RNAs(ceRNAs)在鸡骨骼肌生长发育中发挥作用。Li等[54]通过lncRNA-miRNA-gene互作网络分析及细胞活体试验证明lncIRS1通过招募miR-15家族来调控IRS1基因(IGF-1受体底物基因)的表达,激活IGF-1信号通路相关基因的表达,进而促进肌肉纤维肥大并缓解肌肉萎缩进程。c-Myc是调控细胞增殖、分化和胚胎发生的关键转录因子。在鸡成肌细胞中c-Myc可与1 104个lncRNAs的启动子区结合,这些lncRNAs还可以和MyomiRs形成互作。c-Myc控制着 lncRNA lnc-2949和lnc-1369的转录,而lnc-2949和lnc-1369是MyomiRs的分子海绵,通过吸附肌肉分化相关miRNA,影响成肌细胞的增殖分化(图2)[55]。其次lncRNAs也可以通过编码微肽调节骨骼肌生长发育。在细胞核与细胞质中同时存在的lncRNA-Six1,不但在胸肌组织中高表达,而且可编码产生一个相对分子质量约7 260的微肽,该微肽顺式调控Six1基因、促进骨骼肌细胞增殖的生物学功能,影响鸡肌肉生长[56]。进一步研究还发现miR-1611可与lncRNA-Six1形成内源性竞争,从而调控Six1基因蛋白表达,影响成肌细胞增殖分化及肌纤维类型转化[57]。
|
图 2 c-Myc通过调控靶基因、miRNA和lncRNA影响成肌细胞增殖、分化和肌纤维肥大的作用机制 Fig. 2 Model of c-Myc affecting myoblast proliferation, differentiation and muscle fibre hypertrophy by regulating the expression of its target genes, miRNAs and lncRNAs |
circRNAs是一类广泛存在于动植物细胞、病毒、单细胞真核生物,对基因表达具有调控作用的环形RNAs分子。与线性RNAs分子相比,circRNAs的5′和3′端可通过共价键连接在一起,形成封闭环状结构,而且不易受RNA外切酶影响,表达更加稳定[58-59]。由于circRNAs可以从基因组任意区域产生,因此其长度具有多样性[60]。
20世纪70年代在RNA病毒中首次发现circRNAs[61]。但由于低丰度,功能未知等原因,circRNAs一度不被重视。随着测序技术的不断成熟,研究人员发现circRNAs并不是RNA剪接发生错误形成的垃圾产物,相反在细胞中大量存在,并可以通过不同的途径调节机体平衡,维持机体正常生长发育。此外随着研究的深入,发现存在内部核糖体进入位点(Internal ribosome entry sites,IRES)、开放阅读框(Open reading frame,ORF)时,circRNAs可以翻译有功能的肽。但是与线性mRNAs相比,circRNAs的潜在翻译能力较低[62]。研究还显示circRNAs还具有其他功能,充当miRNAs海绵,作为蛋白海绵和参与转录后调控等[63-65],其中circRNAs作为miRNAs海绵是目前研究最为广泛的作用机制,circRNAs通过吸附miRNAs降低miRNAs和靶mRNAs的结合,间接影响mRNA的翻译。
3.2 circRNAs的研究进展circRNAs是肌肉生长调控网络的新成员,在骨骼肌发育过程中具有独一无二的作用。通过RNA测序技术,Ouyang等[15]在E11、E16和D1 3个时期杏花鸡腿肌中鉴定出13 377个circRNAs。circRNAs作用机制多元,目前在鸡胚骨骼肌发育过程中,circRNAs主要通过作为miRNAs的分子海绵调节成肌细胞的增殖和分化,从而影响骨骼肌的生长发育。在筛选到的多个差异表达的circRNAs中,对鸡胚胎期高表达的circRBFOX2.2-3和circRBFOX2.2-4深入研究,发现它们分别由RBFOX2基因的外显子2-3和外显子2-4环化而成,都可以充当miRNA海绵,通过吸附miR-1a和miR-206调控肌肉细胞的增殖[15]。尽管在其他动物细胞中miR-206对细胞增殖的抑制作用已被广泛报道,但circRNA以miR-206作为中间靶标调控肌肉生长发育的作用机制还是首次报道。来源于SVIL基因外显子6-14的circSVIL在鸡胚发育后期的骨骼肌中高表达,circSVIL与miR-203的5′端种子序列完全配对,充当miR-203的分子海绵调控c-JUN和MEF2C基因表达,从而间接调节成肌细胞增殖和分化过程[66]。来源于FGFR2基因外显子3-6的circFGFR2可与miR-133a-5p和miR-29-5p结合,通过和miR-133a-5p、miR-29-5的相互作用,circFGFR2可调节骨骼肌细胞的增殖分化[67]。同样,研究还发现circHIPK3可充当miR-30a-3p的分子海绵,促进鸡胚骨骼肌细胞的增殖与分化[68]。
4 结论与展望随着研究的深入,科学家们发现编码蛋白质的核苷酸序列仅占基因组的1.5%,它们并不足以完整地解释复杂的生命活动。约98%的基因组DNA都可以被转录成RNA,但绝大多数是非编码的RNA分子。骨骼肌生长发育的调控是一个高度复杂的分子网络,除了有编码功能的基因参与外,ncRNAs等表观遗传调控也扮演了重要角色。目前,家禽骨骼肌发育过程中ncRNAs的研究仍停留在初始阶段。大量新的ncRNAs被鉴定发现,但仅有少量ncRNAs的功能和作用机制被研究清楚。其主要原因有:1)ncRNAs的表达丰度一般比较低,增加了研究其功能机制的难度;2)ncRNAs在物种之间的保守性不强,不同物种之间作用机理也是不同的;3)对家禽动物而言,缺乏专门的ncRNA数据库,ncRNA命名没有统一的标准,缺乏专门的生物信息分析工具。
因此,未来阶段对家禽ncRNAs的研究,应该1)建立专门的数据库,记录ncRNAs的表达丰度、时空表达特异性、组织表达特异性等数据;2)统一ncRNAs命名标准;3)建立针对性强的ncRNAs生物信息学分析工具。与此同时,结合荧光原位杂交、RNA干扰和CRISPR/Cas9介导基因编辑等技术,研究清楚ncRNAs在家禽骨骼肌生长发育中的调控机制,构建基因、miRNAs、lncRNAs和circRNAs之间的互作网络,为促进家禽生长发育提供重要的理论依据。
| [1] |
杨宁. 家禽业的核心将是保持高效优势[J]. 北方牧业, 2018(15): 7. (  0) 0) |
| [2] |
GÜLLER I, RUSSELL A P. MicroRNAs in skeletal muscle: Their role and regulation in development, disease and function[J]. J Physiol, 2010, 588(21): 4075-4087. (  0) 0) |
| [3] |
PEARSON A M. Muscle growth and exercise[J]. Crit Rev Food Sci Nutr, 1990, 29(3): 167-196. DOI:10.1080/10408399009527522 (  0) 0) |
| [4] |
FENG Y, CAO J H, LI X Y, et al. Inhibition of miR-214 expression represses proliferation and differentiation of C2C12 myoblasts[J]. Cell Biochem Funct, 2011, 29(5): 378-383. DOI:10.1002/cbf.1760 (  0) 0) |
| [5] |
SCHIAFFINO S, SANDRI M, MURGIA M. Activity-dependent signaling pathways controlling muscle diversity and plasticity[J]. Physiology (Bethesda), 2007, 22: 269-278. (  0) 0) |
| [6] |
ZANOU N, GAILLY P. Skeletal muscle hypertrophy and regeneration: Interplay between the myogenic regulatory factors (MRFs) and insulin-like growth factors (IGFs) pathways[J]. Cell Mol Life Sci, 2013, 70(21): 4117-4130. DOI:10.1007/s00018-013-1330-4 (  0) 0) |
| [7] |
RULLMAN E, FERNANDEZ-GONZALO R, MEKJAVIĆ I B, et al. MEF2 as upstream regulator of the transcriptome signature in human skeletal muscle during unloading[J]. Am J Physiol Regul Integr Comp Physiol, 2018, 315(4): R799-R809. DOI:10.1152/ajpregu.00452.2017 (  0) 0) |
| [8] |
BRAUN T, GAUTEL M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis[J]. Nat Rev Mol Cell Biol, 2011, 12(6): 349-361. DOI:10.1038/nrm3118 (  0) 0) |
| [9] |
PERRY R L S, RUDNICK M A. Molecular mechanisms regulating myogenic determination and differentiation[J]. Front Biosci, 2000, 5: D750-D767. DOI:10.2741/A548 (  0) 0) |
| [10] |
GAO P F, GUO X H, DU M, et al. LncRNA profiling of skeletal muscles in Large White pigs and Mashen pigs during development[J]. J Anim Sci, 2017, 95(10): 4239-4250. DOI:10.2527/jas2016.1297 (  0) 0) |
| [11] |
CAO Y, YOU S, YAO Y, et al. Expression profiles of circular RNAs in sheep skeletal muscle[J]. Asian-Australas J Anim Sci, 2018, 31(10): 1550-1557. DOI:10.5713/ajas.17.0563 (  0) 0) |
| [12] |
SHI L, ZHOU B, LI P, et al. MicroRNA-128 targets myostatin at coding domain sequence to regulate myoblasts in skeletal muscle development[J]. Cell Signal, 2015, 27(9): 1895-1904. DOI:10.1016/j.cellsig.2015.05.001 (  0) 0) |
| [13] |
DINGER M E, PANG K C, MERCER T R, et al. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities[J]. PloS Comput Biol, 2008, 4(11). doi: 10.1371/journal.pcbi.1000176.
(  0) 0) |
| [14] |
CESANA M, CACCHIARELLI D, LEGNINI I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA[J]. Cell, 2011, 147(2): 358-369. DOI:10.1016/j.cell.2011.09.028 (  0) 0) |
| [15] |
OUYANG H, CHEN X, WANG Z, et al. Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens[J]. DNA Res, 2018, 25(1): 71-86. DOI:10.1093/dnares/dsx039 (  0) 0) |
| [16] |
BALLARINO M, MORLANDO M, FATICA A, et al. Non-coding RNAs in muscle differentiation and musculoskeletal disease[J]. J Clin Invest, 2016, 126(6): 2021-2030. DOI:10.1172/JCI84419 (  0) 0) |
| [17] |
HAYES J, PERUZZI P P, LAWLER S. MicroRNAs in cancer: Biomarkers, functions and therapy[J]. Trends Mol Med, 2014, 20(8): 460-469. DOI:10.1016/j.molmed.2014.06.005 (  0) 0) |
| [18] |
LUO W, NIE Q, ZHANG X. MicroRNAs involved in skeletal muscle differentiation[J]. J Genet Genomics, 2013, 40(3): 107-116. DOI:10.1016/j.jgg.2013.02.002 (  0) 0) |
| [19] |
SAUNDERS M A, LIANG H, LI W H. Human polymorphism at microRNAs and microRNA target sites[J]. Proc Natl Acad Sci USA, 2007, 104(9): 3300-3305. DOI:10.1073/pnas.0611347104 (  0) 0) |
| [20] |
LAI E C. Micro RNAs are complementary to 3′UTR sequence motifs that mediate negative post-transcriptional regulation[J]. Nat Genet, 2002, 30(4): 363-364. DOI:10.1038/ng865 (  0) 0) |
| [21] |
SEOK H, HAM J, JANG E S, et al. MicroRNA target recognition: Insights from transcriptome-wide non-canonical interactions[J]. Mol Cells, 2016, 39(5): 375-381. DOI:10.14348/molcells.2016.0013 (  0) 0) |
| [22] |
LIU G, ZHANG R, XU J, et al. Functional conservation of both CDS- and 3′-UTR-located MicroRNA binding sites between species[J]. Mol Biol Evol, 2015, 32(3): 623-628. (  0) 0) |
| [23] |
KREK A, GRÜN D, POY M N, et al. Combinatorial microRNA target predictions[J]. Nat Genet, 2005, 37(5): 495-500. DOI:10.1038/ng1536 (  0) 0) |
| [24] |
GIRAL H, KRATZER A, LANDMESSER U. MicroRNAs in lipid metabolism and atherosclerosis[J]. Best Pract Res Clin Endocrinol Metab, 2016, 30(5): 665-676. DOI:10.1016/j.beem.2016.11.010 (  0) 0) |
| [25] |
GROSS N, KROPP J, KHATIB H. MicroRNA signaling in embryo development[J]. Biology, 2017, 6(3). doi: 10.3390/biology6030034.
(  0) 0) |
| [26] |
贾新正. 快慢型肉鸡miRNA的表达谱分析[D]. 广州: 华南农业大学, 2010.
(  0) 0) |
| [27] |
WANG X G, YU J F, ZHANG Y, et al. Identification and characterization of microRNA from chicken adipose tissue and skeletal muscle[J]. Poult Sci, 2012, 91(1): 139-149. DOI:10.3382/ps.2011-01656 (  0) 0) |
| [28] |
LIN S, LI H, MU H, et al. Let-7b regulates the expression of the growth hormone receptor gene in deletion-type dwarf chickens[J]. BMC Genomics, 2012, 13. doi: 10.1186/1471-2164-13-306.
(  0) 0) |
| [29] |
WANG X G, SHAO F, GONG D Q, et al. miR-133a targets BIRC5 to regulate its gene expression in chicken[J]. Scientia Agricultura Sinica, 2013, 46(7): 1441-1447. (  0) 0) |
| [30] |
OUYANG H, HE X, LI G, et al. Deep sequencing analysis of miRNA expression in breast muscle of fast-growing and slow-growing broilers[J]. Int J Mol Sci, 2015, 16(7): 16242-16262. DOI:10.3390/ijms160716242 (  0) 0) |
| [31] |
LUO W, WU H, YE Y, et al. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation[J]. Cell Death Dis, 2014, 5. doi: 10.1038/cddis.2014.289.
(  0) 0) |
| [32] |
WANG Z, OUYANG H, CHEN X, et al. Gga-miR-205a affecting myoblast proliferation and differentiation by targeting CDH11[J]. Front Genet, 2018, 9. doi: 10.3389/fgene.2018.00414.
(  0) 0) |
| [33] |
TOWNLEY-TILSON W H D, CALLIS T E, WANG D Z. MicroRNAs 1, 133, and 206: Critical factors of skeletal and cardiac muscle development, function, and disease[J]. Int J Biochem Cell Biol, 2010, 42(8): 1252-1255. DOI:10.1016/j.biocel.2009.03.002 (  0) 0) |
| [34] |
JIA X, LIN H, ABDALLA B A, et al. Characterization of miR-206 promoter and its association with birthweight in chicken[J]. Int J Mol Sci, 2016, 17(4). doi: 10.3390/ijms17040559
(  0) 0) |
| [35] |
LI G, LUO W, ABDALLA B A, et al. miRNA-223 upregulated by MYOD inhibits myoblast proliferation by repressing IGF2 and facilitates myoblast differentiation by inhibiting ZEB1[J]. Cell Death Dis, 2017, 8. doi: 10.1038/cddis.2017.479.
(  0) 0) |
| [36] |
GUO L, HUANG W, CHEN B, et al. Gga-mir-133a-3p regulates myoblasts proliferation and differentiation by targeting PRRX1[J]. Front Genet, 2018, 9. doi: 10.3389/fgene.2018.00577.
(  0) 0) |
| [37] |
WANG Z, ZHANG X, LI Z, et al. MiR-34b-5p mediates the proliferation and differentiation of myoblasts by targeting IGFBP2[J]. Cells, 2019, 8(4). doi: org/10.3390/cells8040360.
(  0) 0) |
| [38] |
WANG J, HELIN K, JIN P, et al. Inhibition of in vitro myogenic differentiation by cellular transcription factor E2F1
[J]. Cell Growth Differ, 1995, 6(10): 1299-1306. (  0) 0) |
| [39] |
LUO W, LI G, YI Z, et al. E2F1-miR-20a-5p/20b-5p auto-regulatory feedback loop involved in myoblast proliferation and differentiation[J]. Sci Rep. 2016, 6. doi: 10.1038/srep27904.
(  0) 0) |
| [40] |
JIA X, OUYANG H, ABDALLA B A, et al. miR-16 controls myoblast proliferation and apoptosis through directly suppressing Bcl2 and FOXO1 activities[J]. Biochim Biophys Acta Gene Regul Mech, 2017, 1860(6): 674-684. DOI:10.1016/j.bbagrm.2017.02.010 (  0) 0) |
| [41] |
JIA X, LIN H, NIE Q, et al. A short insertion mutation disrupts genesis of miR-16 and causes increased body weight in domesticated chicken[J]. Sci Rep, 2016, 6. doi: 10.1038/srep36433.
(  0) 0) |
| [42] |
YANG Y L, LOH K S, LIOU B Y, et al. SESN-1 is a positive regulator of lifespan in Caenorhabditis elegans
[J]. Exp Gerontol, 2013, 48(3): 371-379. DOI:10.1016/j.exger.2012.12.011 (  0) 0) |
| [43] |
EL HUSSEINI N, HALES B F. The roles of P53 and its family proteins, P63 and P73, in the DNA damage stress response in organogenesis stage mouse embryos[J]. Toxicol Sci, 2018, 162(2): 439-449. DOI:10.1093/toxsci/kfx270 (  0) 0) |
| [44] |
CAI B, MA M, CHEN B, et al. MiR-16-5p targets SESN1 to regulate the p53 signaling pathway, affecting myoblast proliferation and apoptosis, and is involved in myoblast differentiation[J]. Cell Death Dis, 2018, 9. doi: 10.1038/s41419-018-0403-6.
(  0) 0) |
| [45] |
CABILI M N, TRAPNELL C, GOFF L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses[J]. Genes Dev, 2011, 25(18): 1915-1927. (  0) 0) |
| [46] |
OKAZAKI Y, FURUNO M, KASUKAWA T, et al. Analysis of the mouse transcriptome based on functional annotation of 60, 770 full-length cDNAs[J]. Nature, 2002, 420(6915): 563-573. DOI:10.1038/nature01266 (  0) 0) |
| [47] |
WILUSZ J E, SUNWOO H, SPECTOR D L. Long noncoding RNAs: Functional surprises from the RNA world[J]. Genes Dev, 2009, 23(13): 1494-1504. DOI:10.1101/gad.1800909 (  0) 0) |
| [48] |
SANLI I, LALEVÉE S, CAMMISA M, et al. Meg3 non-coding RNA expression controls imprinting by preventing transcriptional upregulation in cis
[J]. Cell Rep, 2018, 23(2): 337-348. DOI:10.1016/j.celrep.2018.03.044 (  0) 0) |
| [49] |
KALLEN A N, ZHOU X B, XU J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs[J]. Mol Cell, 2013, 52(1): 101-112. DOI:10.1016/j.molcel.2013.08.027 (  0) 0) |
| [50] |
ZHOU L, SUN K, ZHAO Y, et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1[J]. Nat Commun, 2015, 6. doi: 10.1038/ncomms10026.
(  0) 0) |
| [51] |
LI T, WANG S, WU R, et al. Identification of long non-protein coding RNAs in chicken skeletal muscle using next generation sequencing[J]. Genomics, 2012, 99(5): 292-298. DOI:10.1016/j.ygeno.2012.02.003 (  0) 0) |
| [52] |
LI Z, OUYANG H, ZHENG M, et al. Integrated analysis of long non-coding RNAs (lncRNAs) and mRNA expression profiles reveals the potential role of lncRNAs in skeletal muscle development of the chicken[J]. Front Physiol, 2017, 7. doi: 10.3389/fphys.2016.00687.
(  0) 0) |
| [53] |
OUYANG H, WANG Z, CHEN X, et al. Proteomic analysis of chicken skeletal muscle during embryonic development[J]. Front Physiol, 2017, 8. doi: 10.3389/fphys.2017.00281.
(  0) 0) |
| [54] |
LI Z, CAI B, ABDALLA B A, et al. LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway[J]. J Cachexia Sarcopenia Muscle, 2019, 10(2): 391-410. DOI:10.1002/jcsm.v10.2 (  0) 0) |
| [55] |
LUO W, CHEN J, LI L, et al. c-Myc inhibits myoblast differentiation and promotes myoblast proliferation and muscle fibre hypertrophy by regulating the expression of its target genes, miRNAs and lincRNAs[J]. Cell Death Differ, 2019, 26(3): 426-442. DOI:10.1038/s41418-018-0129-0 (  0) 0) |
| [56] |
CAI B, LI Z, MA M, et al. LncRNA-Six1 encodes a micropeptide to activate Six1 in cis and is involved in cell proliferation and muscle growth[J]. Front Physiol, 2017, 8. doi: 10.3389/fphys.2017.00230.
(  0) 0) |
| [57] |
MA M, CAI B, JIANG L, et al. LncRNA-Six1 is a target of miR-1611 that functions as a ceRNA to regulate Six1 protein expression and fiber type switching in chicken myogenesis[J]. Cells, 2018, 7(12). doi: 10.3390/cells7120243.
(  0) 0) |
| [58] |
DANAN M, SCHWARTZ S, EDELHEIT S, et al. Transcriptome-wide discovery of circular RNAs in archaea[J]. Nucleic Acids Res, 2012, 40(7): 3131-3142. DOI:10.1093/nar/gkr1009 (  0) 0) |
| [59] |
CHEN L L, YANG L. Regulation of circRNA biogenesis[J]. RNA Biol, 2015, 12(4): 381-388. DOI:10.1080/15476286.2015.1020271 (  0) 0) |
| [60] |
ENUKA Y, LAURIOLA M, FELDMAN M E, et al. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor[J]. Nucleic Acids Res, 2016, 44(3): 1370-1383. DOI:10.1093/nar/gkv1367 (  0) 0) |
| [61] |
WU Q, WANG Y, CAO M, et al. Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm[J]. Proc Natl Acad Sci USA, 2012, 109(10): 3938-3943. DOI:10.1073/pnas.1117815109 (  0) 0) |
| [62] |
LEGNINI I, DI TIMOTEO G, ROSSI F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis[J]. Mol Cell, 2017, 66(1): 22-37. DOI:10.1016/j.molcel.2017.02.017 (  0) 0) |
| [63] |
HANSEN T B, JENSEN T I, CLAUSEN B H, et al. Natural RNA circles function as efficient microRNA sponges[J]. Nature, 2013, 495(7441): 384-388. DOI:10.1038/nature11993 (  0) 0) |
| [64] |
MEMCZAK S, JENS M, ELEFSINIOTI A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency[J]. Nature, 2013, 495(7441): 333-338. DOI:10.1038/nature11928 (  0) 0) |
| [65] |
DU W W, YANG W, LIU E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2[J]. Nucleic Acids Res, 2016, 44(6): 2846-2858. DOI:10.1093/nar/gkw027 (  0) 0) |
| [66] |
OUYANG H, CHEN X, LI W, et al. Circular RNA circSVIL promotes myoblast proliferation and differentiation by sponging miR-203 in chicken[J]. Front Genet, 2018, 9. doi: 10.3389/fgene.2018.00172.
(  0) 0) |
| [67] |
CHEN X, OUYANG H, WANG Z, et al. A novel circular RNA generated by FGFR2 gene promotes myoblast proliferation and differentiation by sponging miR-133a-5p and miR-29b-1-5p[J]. Cells, 2018, 7(11). doi: 10.3390/cells7110199.
(  0) 0) |
| [68] |
CHEN B, YU J, GUO L, et al. Circular RNA circHIPK3 promotes the proliferation and differentiation of chicken myoblast cells by sponging miR-30a-3p[J]. Cells, 2019, 8(2). doi: 10.3390/cells8020177.
(  0) 0) |
 2019, Vol. 40
2019, Vol. 40

