2. 广州国家现代农业产业科技创新中心,广东 广州 510520;
3. 华南农业大学 亚热带农业生物资源保护与利用国家重点实验室,广东 广州 510642;
4. 肯塔基大学 昆虫学系,肯塔基州 列克星敦市 40546
2. National Modern Agricultural Industry Science and Technology Innovation Center of Guangzhou, Guangzhou 510520, China;
3. State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, South China Agricultural University, Guangzhou 510642, China;
4. Department of Entomology, University of Kentucky, Lexington 40546, USA
Henosepilachna vigintioctopunctata (Fabricius) and Henosepilachna vigintioctomaculata (Motschulsky) (Coleoptera: Coccinellidae) have similar morphological characteristics and host ranges and are serious agricultural pests in Asian countries[1-5]. They both feed on the leaves of solanaceous plants, and young stems, petals, sepals, and fruits will also be harmed[6-7]. Although they have becoming increasingly serious agricultural pests in China, Asia and globally[3,5,8-10], there is still no efficient way to differentiate them, due to the subtle differences in morphological characteristics. In traditional morphological taxonomy of insects, well-preserved adult specimens can usually be differentiated based on the difference in the lines on the pronotum and the black spot arrangement on the base of elytra[11]. However, to distinguish these two insects accurately, additional factors need to be considered, including the structure of the male and female genitals, and the longitudinal crack on the sternum[12]. This requires professional taxonomic knowledge, sufficient experience and examination at a specified life stage. To develop targeted management practices for the control of H. vigintioctopunctata and H. vigintioctomaculata, finding a method to distinguish them rapidly and accurately is imperative.
Molecular techniques to identify eukaryotes rapidly and effectively with only a small fragment of DNA have come into their own in recent years. Random amplified polymorphic DNA (RAPD) markers have been used to distinguish aphids, but the change in reaction conditions makes it unstable and easy to produce variation[13]. Previous studies have shown that RAPD markers can be converted into sequence characterized amplified region (SCAR) markers by designing specific primers based on DNA sequences. SCAR is widely used in insect species identification[14]. In 2002, scientists raised the concept of DNA taxonomy[15]. They elucidated that the identification difficulties produced by high degrees of similarity between phylogenetically close species would be solved, and DNA taxonomy would become a reliable tool for species identification with DNA sequences as the universal reference standard in taxonomy[15].
Based on the mitochondrial cytochrome oxidase I gene (mtCOI), species-specific (SS) PCR identifies the species with specific primers and PCR programs. In recent years, studies have reported that SS-mtCOI PCR has a high sensitivity and strong specificity in the identification of pests. Moreover, it is not restricted by the developmental stage and integrity of specimens, indicating a strong application in quarantine, monitoring, and pest management[16]. For examples, the citrus mealybug Planococcus citri Risso, the obscure mealybug Pseudococcus viburni Signoret, and the comstock mealybug Pseudococcus comstocki Kuwana can be distinguished effectively by SS-mtCOI primers[17]. SS-mtCOI primers were designed to identify six similar stored-product pests[18]. SS-mtCOI primers were used for the identification assay of Sirex noctilio[19]. All of these studies indicate that SS-mtCOI PCR technology is an efficient method to identify phylogenetically close species.
In this research, we established the SS-mtCOI PCR program based on analyzing mtCOI sequences and designing SS-mtCOI primers with the aim for differentiating H. vigintioctopunctata and H. vigintioctomaculata. The sensitivity of the primers was investigated under different DNA concentrations, and the efficiency was validated at the egg, and 1st instar stages of insects. Furthermore, these primers were used to identify H. vigintioctopunctata field populations which were collected from six provinces.
2 Materials and Methods 2.1 InsectsHenosepilachna vigintioctopunctata adults were collected from Solanum nigrum (L.) on the campus of South China Agricultural University in April 2018, Guangzhou City, Guangdong Province, China, as described previously[5]. H. vigintioctomaculata adults were collected from potato leaves in the practice teaching base station of Hebei North University in May 2018, Zhangjiakou City, Hebei Province, China. Ladybeetles were identified by a specialist entomologist. Each stage of the two ladybeetles were photographed with an Olympus stereomicroscope (SZX16, Olympus, Tokyo, Japan) connected to a EOS 5D Mark IV Camera (Canon, Tokyo, Japan) (Fig. 1), we can see that it is very hard to distinguish them based on the morphological characteristics.

|
Fig. 1 The representative morphological characteristics of Henosepilachna vigintioctopunctata and H. vigintioctomaculata at each developmental stage |
Genomic DNA was extracted from four H. vigintioctopunctata and H. vigintioctomaculata adult individuals using a commercial TIANamp Genomic DNA kit (TIANGEN, Beijing, China) according to the manufacturer’s protocol. The DNA concentration was determined using a NanoDrop OneC spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). The DNA samples were stored at −20℃ until used for PCR analysis. A pair of universal forward LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and reverse HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) primers were used for the mtCOI amplification. PCR amplifications and parameters were performed according to our previous study[20]. PCR amplifications were performed in 20.0 μL reactions containing 10.0 μL MasterMix with loading dye, 7.0 μL sterilized distilled water, 1.0 μL DNA template, and 1.0 μL forward and reverse primers (10 μmol/L). The PCR parameters were as follows: Denaturation at 94 ℃for 3 min, followed by 35 cycles (94 ℃ for 30 s, 55 ℃ for 45 s, 72 ℃ for 1 min), and a final extension at 72 ℃ for 10 min. The amplified DNA fragments were resolved on a 10 g/L agarose gel (1 × Tris Acetate-EDTA buffer), stained with Goldview I, and photographed. DNA purification was conducted using a DNA purification kit (TIANGEN, Beijing, China). Three PCR amplicons for each insect species were sequenced by one directional sequencing with forward primer LCO1490 by TSINGKE Biological Technology (TSINGKE, Guangzhou, China). The pairwise sequence alignments of mtCOI genes were carried out between H. vigintioctopunctata and H. vigintioctomaculata using Geneious 7.0.
2.3 Specificity of the SS-mtCOI primersAfter alignment and analysis of the partial sequences of mtCOI from H. vigintioctopunctata and H. vigintioctomaculata, SS-mtCOI primers were designed for each species. SS-mtCOI primers for H. vigintioctopunctata were forward primer Hvp-F (5′-GGCTTTCCCTCGACTAAAT-3′) and reverse primer Hvp-R (5′-ACCTCCTGCAGGGTCA-3′); SS-mtCOI primers for H. vigintioctomaculata were forward primer Hvm-F (5′-TGATATAGCTTTTCCTCGACTTAAC-3′) and reverse primer Hvm-R(5′-TCCTCCAGCTGGATCG-3′). The PCR parameters were as follows: Denaturation at 94 ℃ for 3 min, followed by 35 cycles (94 ℃ for 30 s, 65 ℃ for 45 s, 72 ℃ for 1 min), and a final extension at 72 ℃ for 10 min. DNA from H. vigintioctopunctata and H. vigintioctomaculata was used as the template to assay the specificity of the primers.
2.4 Sensitivity of the SS-mtCOI PCR assayTo assess the sensitivity of the SS-mtCOI PCR assay, the threshold of the species-specific primers was determined by using double gradient dilutions of DNA from a whole adult of the two ladybeetles. The PCR programs and parameters were conducted as described in “2.3”.
2.5 Verification of the SS-mtCOI primers in the egg and 1st instarGenomic DNA of single egg or 1st instar was extracted from H. vigintioctopunctata and H. vigintioctomaculata individuals as described in “2.2”. PCR programs and parameters were used as described in “2.3”.
2.6 Verification of the SS-mtCOI primers in six field populations of H. vigintioctopunctataSix populations of H. vigintioctopunctata were collected from six provinces to verify the specificity of Hvp and Hvm primers, including Guangzhou City of Guangdong Province, Jingzhou City of Hubei Province, Hangzhou City of Zhejiang Province, Nanjing City of Jiangsu Province, Heze City of Shandong Province, and Beijing City. Genomic DNA of H. vigintioctopunctata individuals were extracted as described above for PCR detection, and three individuals were tested for each population.
3 Results and analysis 3.1 mtCOI sequences analysis between H. vigintioctopunctata and H. vigintioctomaculataUsing DNA from H. vigintioctopunctata and H. vigintioctomaculata as the template, PCR amplification was performed with the universal mtCOI gene primers LCO1490/HC02198 in four replications. Gel electrophoresis showed that PCR amplified a clear fragment in all the samples (Fig. 2A). Pairwise comparisons indicated that H. vigintioctopunctata and H. vigintioctomaculata mtCOI partial sequences (685 bp) have 88.2% similarity (Fig. 2B).

|
Fig. 2 PCR amplicons from Henosepilachna vigintioctopunctata and H. vigintioctomaculata using mtCOI gene universal primers (A) and pairwise comparisons of the mtCOI sequences (B) Marker: DL2000 DNA Marker;1–4: H. vigintioctopunctata; 5–8: H. vigintioctomaculata |
The specificity of the SS-mtCOI primers (Hvp and Hvm) was tested for H. vigintioctopunctata and H. vigintioctomaculata. Results showed that Hvp and Hvm primers had species-specific amplifications. Specifically, Hvp primers had a 400 bp band in H. vigintioctopunctata (Fig. 3A), while Hvm primers had a 406 bp band in H. vigintioctomaculata (Fig. 3B).
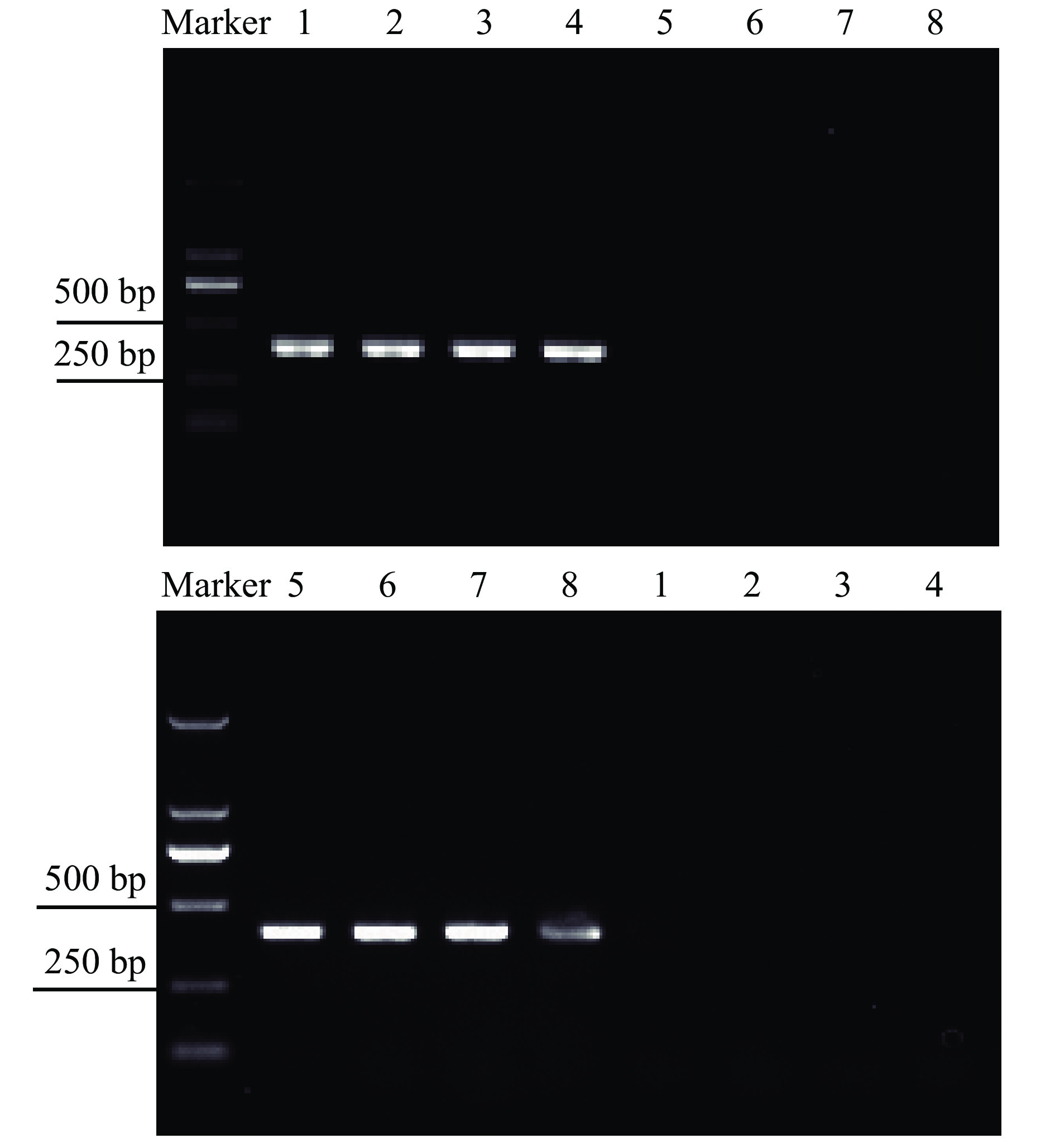
|
Fig. 3 Amplification patterns of mtCOI of Henosepilachna vigintioctopunctata and H. vigintioctomaculata with Hvp primers (A) and Hvm primers (B) Marker: DL2000 DNA Marker; 1–4: H. vigintioctopunctata; 5–8: H. vigintioctomaculata |
PCR was performed with double gradient diluted template DNA from H. vigintioctopunctata and H. vigintioctomaculata. Gel electrophoresis showed that an amplification band was detected when the DNA from H. vigintioctopunctata was diluted 32 times (3.13 mg/L, Fig. 4A), and when the DNA from H. vigintioctomaculata was diluted 64 times (2.43 mg/L, Fig. 4B).
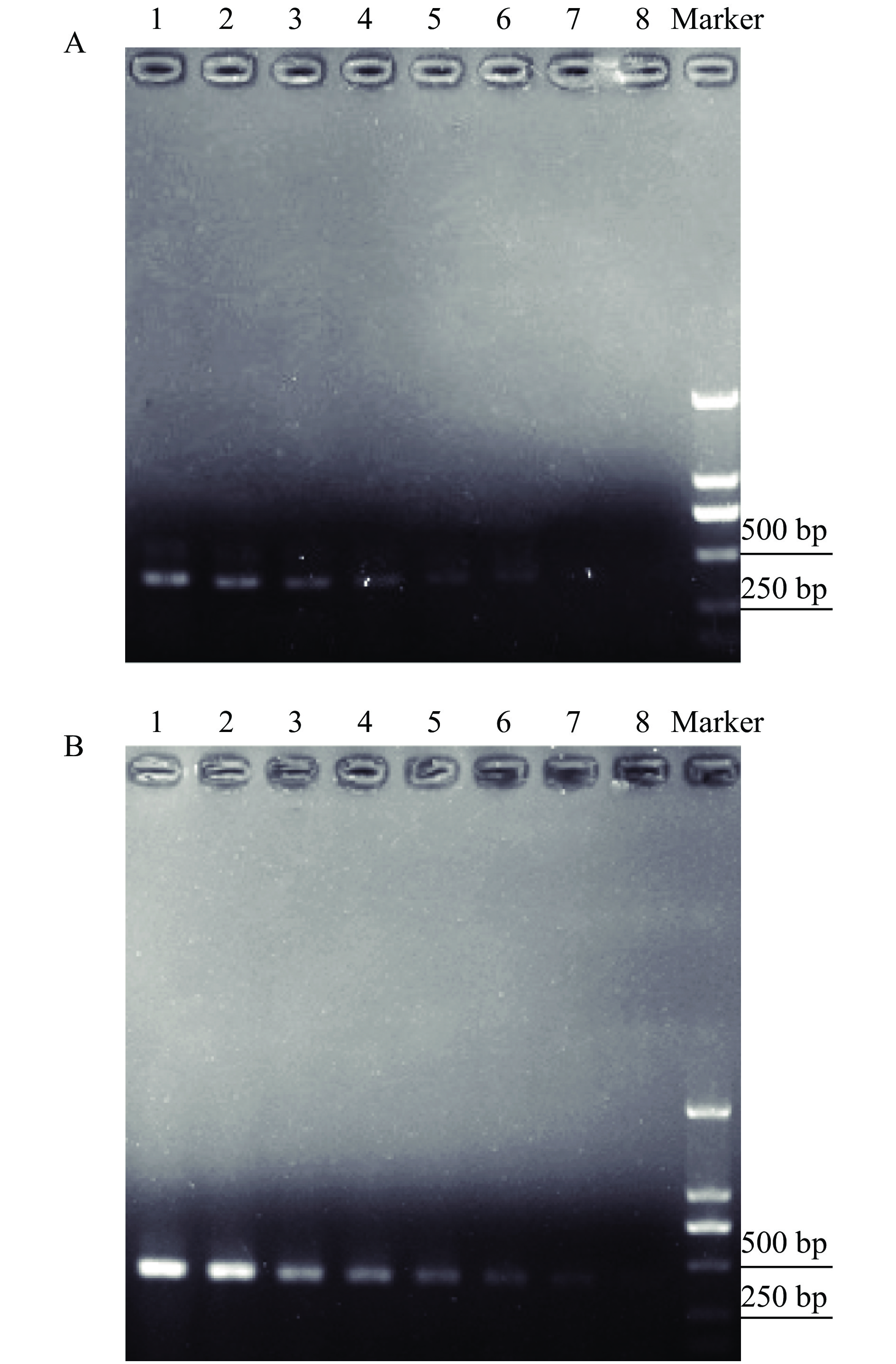
|
Fig. 4 Gel electrophoresis of PCR products amplified from double diluted Henosepilachna vigintioctopunctata template DNA with Hvp primers (A) and H. vigintioctomaculata template DNA with Hvm primers (B) Marker: DL2000 DNA Marker; For figure A, 1–8: Dilution of 100.00, 50.00, 25.00, 12.50, 6.25, 3.13, 1.56, and 0.78 mg/L respectively; For figure B, 1–8: Dilution of 150.00, 75.00, 37.50, 18.75, 9.38, 4.69, 2.43, and 1.17 mg/L respectively |
PCR was performed with single egg or single 1st instar template DNA from H. vigintioctopunctata and H. vigintioctomaculata. Gel electrophoresis showed that Hvp and Hvm primers could be used to distinguish H. vigintioctopunctata and H. vigintioctomaculata at the egg or the 1st instar stage (Fig. 5).
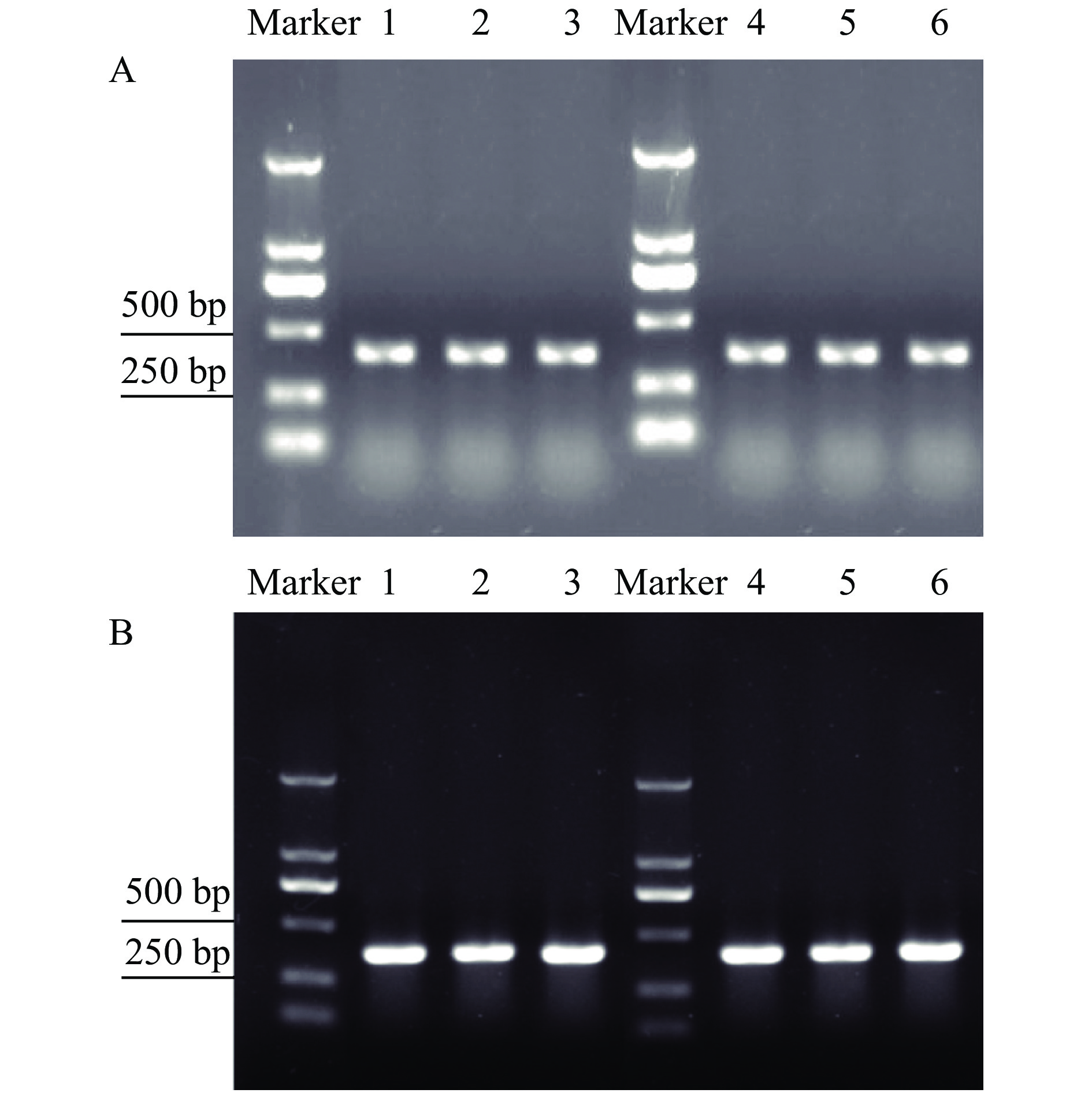
|
Fig. 5 Gel electrophoresis of PCR products amplified from the egg and 1st instar of Henosepilachna vigintioctopunctata template DNA with Hvp primers (A) and H. vigintioctomaculata template DNA with Hvm primers (B) Marker:DL2000 DNA Marker; 1–3: Egg; 4–6: 1st instar |
Gel electrophoresis showed that Hvp primers had specific amplicons in all the six field populations, whereas Hvm primers had no amplicon (Fig. 6).
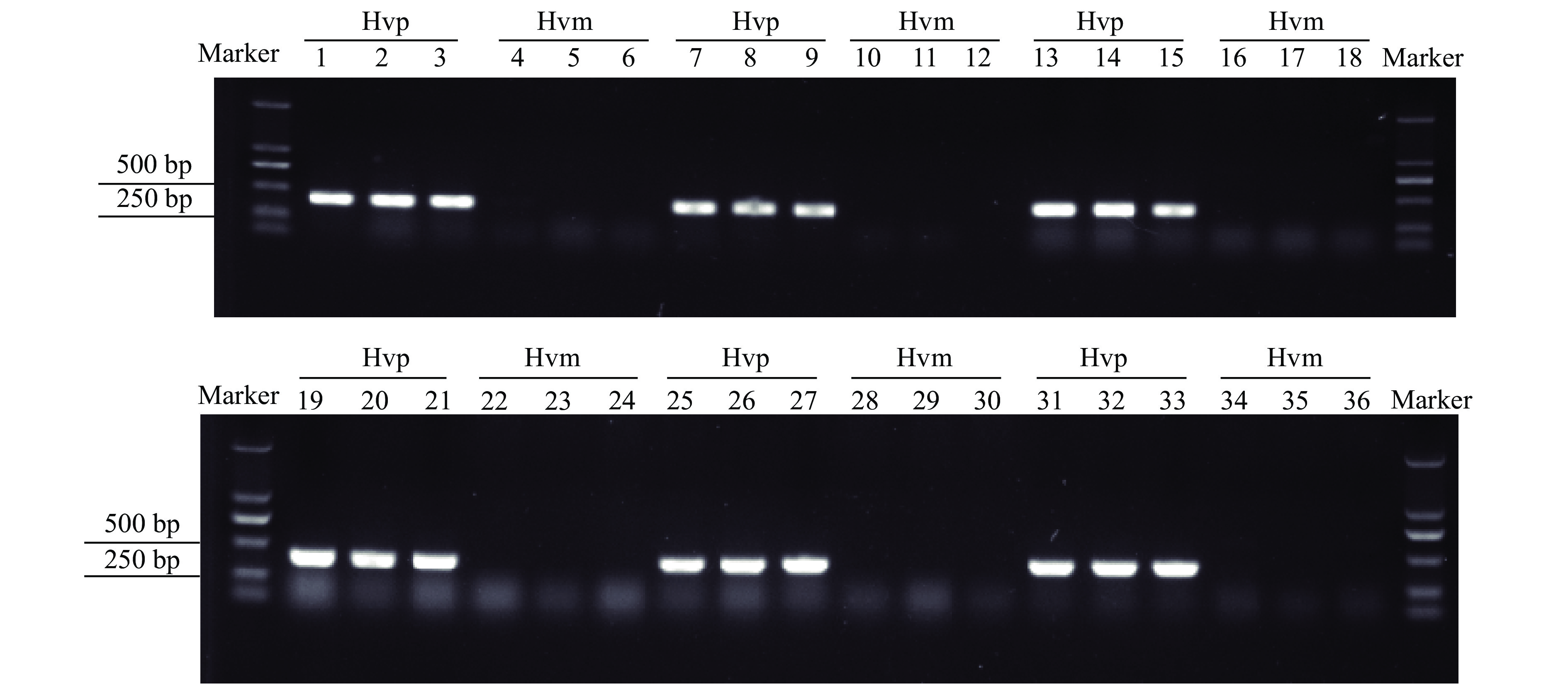
|
Fig. 6 Gel electrophoresis of PCR products amplified from six different populations of Henosepilachna vigintioctopunctata with Hvp and Hvm primers Marker: DL2000 DNA Marker; 1–6: Samples from Guangzhou City, 7–12: Samples from Hangzhou City, 13–18: Samples from Jingzhou City. 19–24: Samples from Nanjing City, 25–30: Samples from Heze City, 31–36: Samples from Beijing City |
In recent years, owing to warming of the climate, development of trade, and expansion of potato planting, food for H. vigintioctopunctata and H. vigintioctomaculata is constantly provided, and the occurrence and damage caused by H. vigintioctopunctata and H. vigintioctomaculata has become much more serious in China[3,21]. With the similar morphological characters, they are difficult to be distinguished through traditional taxonomic methods. Hence, a method to distinguish H. vigintioctopunctata and H. vigintioctomaculata rapidly and accurately is of great significance for insect taxonomy and pest management. Compared with conventional morphological identification[11], the molecular assays are more efficient and sensitive, and not limited by the life stage and specimen quality[16].
The mtCOI gene is one of the conserved, protein-coding genes in the mitochondrial genome, which can be amplified by universal primers, providing a robust DNA barcode for identifying species[22]. SS-mtCOI PCR technology is a promising tool in entomological taxonomy, as it demonstrates high divergence between species[23]. In the current study, the SS-mtCOI PCR assay based on the mtCOI gene sequence analysis was used to distinguish H. vigintioctopunctata and H. vigintioctomaculata rapidly and accurately. The presence of the specificity fragments confirmed that the SS-mtCOI primers were sufficiently species-specific in both laboratory and field populations. Furthermore, these two primers were used in the same PCR programs with low concentrations of DNA and showed that the egg and 1st instar of H. vigintioctopunctata and H. vigintioctomaculata could be distinguished by Hvp and Hvm primers. In view of 88.2% similarity in H. vigintioctopunctata and H. vigintioctomaculata mtCOI partial sequences (685 bp), it is difficult to design specific primers for the PCR analysis to distinguish them. Here, the most important parameter in the PCR programs was the 65 ℃ annealing temperature. When the PCR was conducted with lower annealing temperature, like 55 ℃, Hvp and Hvm primers had no species-specific amplification. This is consistent with previous study using SS-mtCOI primers with 64 ℃ annealing temperature to distinguish the whitefly Bemisia tabaci B and Q cryptic species, which cannot be identified by morphological traits[24].
The DNA detection threshold value was 3.13 mg/L of H. vigintioctopunctata with Hvp primers, and 2.43 mg/L of H. vigintioctomaculata with Hvm primers. In other studies of the same field, for examples, the DNA detection value using SS-mtCOI primers was 0.06 mg/L for Pseudococcus jackbeardsleyi[16], 0.144 mg/L for Phenacoccus manihoti[25], and 0.19 mg/L for Phenacoccus madeirensis[26]. Compared to these primers, the detection sensitivity of Hvp and Hvm primers was lower, however, Hvp and Hvm primers were sensitively enough for differentiating H. vigintioctopunctata and H. vigintioctopunctala even at the egg and 1st instar stages.
Taken together, the SS-mtCOI PCR identification assay can specifically and sensitively distinguish the two ladybeetles in a short time and with low concentrations of DNA. This SS-mtCOI PCR method is an obvious update on traditional identification methods, and will facilitate the precise management of these two kinds of morphological indistinguishable ladybeetles in the field.
| [1] |
RAHAMAN M A, PRODHAN M D H, MAULA A K M. Effect of botanical and synthetic pesticides in controlling Epilachna beetle and the yield of bitter gourd
[J]. International Journal of Sustainable Crop Production, 2008, 3(5): 23-26. (  0) 0) |
| [2] |
ISLAM K, ISLAM M S, FERDOUSI Z. Control of Epilachna vigintioctopuntata Fab. (Coleoptera: Coccinellidae) using some indigenous plant extracts
[J]. Journal of Life and Earth Science, 2011, 6: 75-80. (  0) 0) |
| [3] |
ZHOU L, XIE B G, WANG X P. Population dynamic of Henosepilachna vigintioctopunctata in different host plants in Jianghan Plain
[J]. Northern Horticulture, 2015, 11: 103-105. (  0) 0) |
| [4] |
KOHYAMA T I, MATSUBAYASHI K W, KATAKURA H. Heterospecific sperm reduction in interspecific crosses between two closely related phytophagous ladybird beetles, Henosepilachna vigintioctomaculata and H. pustulosa (Coleoptera: Coccinellidae)
[J]. Entomological Science, 2016, 19: 49-54. DOI:10.1111/ens.12159 (  0) 0) |
| [5] |
LÜ J, CHEN S M, GUO M J, et al. Selection and validation of reference genes for RT-qPCR analysis of the ladybird beetle Henosepilachna vigintioctomaculata
[J]. Frontiers in Physiology, 2018, 9: 1614. DOI:10.3389/fphys.2018.01614 (  0) 0) |
| [6] |
KARTHIKA P, VADIVALAGAN C, KRISHNAVENI N, et al. Contrasting genetic diversity and intra-population polymorphism of the invasive pest Henosepilachna vigintioctopunctata (Coleoptera, Coccinellidae): A DNA barcoding approach
[J]. Journal of Asia-Pacific Entomology, 2017, 20: 23-29. DOI:10.1016/j.aspen.2016.11.011 (  0) 0) |
| [7] |
KARTHIKA P, VADIVALAGAN C, MUTHUSANKAR A, et al. Methyl linolenate as a feeding stimulant for the 28-spotted potato ladybird, Henosepilachna vigintioctopunctata? A molecular docking approach
[J]. Physiological and Molecular Plant Pathology, 2018, 101: 75-84. DOI:10.1016/j.pmpp.2017.01.005 (  0) 0) |
| [8] |
GHOSH S K, SENAPATI S K. Biology and seasonal fluctuation of Henosepilachna vigintioctopunctata Fabr. on brinjal under Terai region of West Bengal
[J]. Indian Journal of Agricultural Research, 2001, 35: 149-154. (  0) 0) |
| [9] |
SHINOGI T, HAMANISHI Y, OTSU Y, et al. Role of induced resistance in interactions of Epilachna vigintioctopunctata with host and non-host plant species
[J]. Plant Science, 2005, 168: 1477-1485. DOI:10.1016/j.plantsci.2005.01.022 (  0) 0) |
| [10] |
VENKATESHA M G. Seasonal occurrence of Henosepilachna vigintioctopunctata (F.) (Coleoptera: Coccinellidae) and its parasitoid on Ashwagandha in India
[J]. Journal of Asia-Pacific Entomology, 2006, 9(3): 265-268. DOI:10.1016/S1226-8615(08)60301-5 (  0) 0) |
| [11] |
ZHANG Y C, LIU H J, ZHENG Z M. Ultrastructure of Henosepilachna vigintioctomaculata and H. vigintioctopunctata
[J]. Chinese Bulletin of Entomology, 2002, 39: 132-135. (  0) 0) |
| [12] |
YU G Y. Identification of “28 stars” ladybug[J]. Chinese Bulletin of Entomology, 2000, 37: 239-242. (  0) 0) |
| [13] |
BLACK W C, DUTEAU N M, PUTERKA G J, et al. Use of the random amplified polymorphic DNA polymerase chain reaction (RAPD-PCR) to detect DNA polymorphisms in aphids (Homoptera: Aphididae)[J]. Bulletin of Entomological Research, 1992, 82: 151-159. DOI:10.1017/S0007485300051671 (  0) 0) |
| [14] |
WANG N, WANG X, ZHANG Y, et al. Establishment and application of SCAR marker for rapid identification of Trialeurodes vaporariorum
[J]. Journal of Environmental Entomology, 2010, 53: 323-330. (  0) 0) |
| [15] |
TAUTZ D, ARCTANDER P, MINELLI A, et al. DNA points the way ahead in taxonomy[J]. Nature, 2002, 418(6897): 479. (  0) 0) |
| [16] |
ZHANG G F, WANG Y S, GUO J Y, et al. Rapid identification of the important quarantine pest Diabrotica virgifera virgifera by SS-COI
[J]. Plant Protection, 2019, 45: 109-115. (  0) 0) |
| [17] |
WANG Y S, ZHOU P, TIAN H, et al. First record of the invasive pest Pseudococcus jackbeardsleyi (Hemiptera: Pseudococcidae) on the Chinese mainland and its rapid identification based on species-specific polymerase chain reaction
[J]. Journal of Economic Entomology, 2018, 111(5): 2120-2128. DOI:10.1093/jee/toy223 (  0) 0) |
| [18] |
ZHANG T, WANG Y, GUO W, et al. DNA barcoding, species-specific PCR and real-time PCR techniques for the identification of six Tribolium pests of stored products[J]. Scientific Reports, 2016, 6: 28494. DOI:10.1038/s41598-016-0001-8 (  0) 0) |
| [19] |
SUN X T, TAO J, REN L L, et al. Identification of Sirex noctilio (Hymenoptera: Siricidae) using a species-specific cytochrome C oxidase subunit I PCR assay
[J]. Journal of Economic Entomology, 2016, 109(3): 1424-1430. DOI:10.1093/jee/tow060 (  0) 0) |
| [20] |
PAN H, CHU D, GE D, et al. Further spread of and domination by Bemisia tabaci (Hemiptera: Aleyrodidae) biotype Q on field crops in China
[J]. Journal of Economic Entomology, 2011, 104(3): 978-985. DOI:10.1603/EC11009 (  0) 0) |
| [21] |
LÜ J, GUO W, GUO M J, et al. Double-stranded RNAs targeting HvRPS18 and HvRPL13 reveal potential targets for pest management of the 28-spotted ladybeetle, Henosepilachna vigintioctopunctata
[J]. Pest Management Science, 2020, 76(8): 2663-2673. DOI:10.1002/ps.5809 (  0) 0) |
| [22] |
HEBERT P D, CYWINSKA A, BALL S L, et al. Biological identifications through DNA barcodes[J]. Proceedings of the Royal Society B: Biological Sciences, 2003, 270(1512): 313-321. DOI:10.1098/rspb.2002.2218 (  0) 0) |
| [23] |
JORDAENS K, SONET G, RICHET R, et al. Identification of forensically important Sarcophaga species (Diptera: Sarcophagidae) using the mitochondrial COI gene
[J]. International Journal of Legal Medicine, 2013, 127(2): 491-504. DOI:10.1007/s00414-012-0767-6 (  0) 0) |
| [24] |
SHATTERS R G, POWELL C A, BOYKIN L M, et al. Improved DNA barcoding method for Bemisia tabaci and related Aleyrodidae: Development of universal and Bemisia tabaci biotype-specific mitochondrial cytochrome c oxidase I polymerase chain reaction primers
[J]. Journal of Economic Entomology, 2009, 102(2): 750-758. DOI:10.1603/029.102.0236 (  0) 0) |
| [25] |
WANG Y S, TIAN H, WAN F H, et al. Species-specific COI primers for rapid identification of a globally significant invasive pest, the cassava mealybug Phenacoccus manihoti Matile-Ferrero
[J]. Journal of Integrative Agriculture, 2019, 18: 1042-1049. DOI:10.1016/S2095-3119(18)62043-X (  0) 0) |
| [26] |
WANG Y S, DAI T M, TIAN H, et al. Phenacoccus madeirensis green (Hemiptera: Pseudococcidae): New geographic records and rapid identification using a species-specific PCR assay
[J]. Crop Protection, 2019, 116: 68-76. DOI:10.1016/j.cropro.2018.10.003 (  0) 0) |
 2022, Vol. 43
2022, Vol. 43


