种子活力与种植业生产关系十分密切。北美官方种子分析家协会(Association of official seed analysts,AOSA)将种子活力定义为:种子活力是指在广泛的田间条件下,决定种子迅速整齐出苗和长成正常幼苗潜在能力的总称。因此,种子活力可以具体表现为:种子发芽和幼苗生长的速度和整齐度;田间表现,包括出苗、生长的速度和整齐度;贮藏、运输后的表现,特别是发芽能力的保持,以及种子耐逆性等[1]。种子活力随着生理成熟而逐渐增加,在收获前后开始下降[2],在种子储藏过程中活力也会逐渐降低[3]。近年来,利用QTL定位、全基因组关联分析,以及各种组学方法鉴定了大量控制作物种子活力相关基因,并在种子活力分子机理研究方面取得了长足发展,主要涉及激素、氨基酸、活性氧(Reactive oxygen species,ROS)、能量代谢、光形态建成等。近来,作物轻简栽培比如直播稻等的广泛应用,亟需生产高活力的作物种子[4]。因此,种子活力已成为种子科学与技术领域研究热点。本文主要以水稻Oryza sativa和拟南芥Arabidopsis thaliana等模式植物为例,重点介绍种子发育成熟过程中活力形成、种子快速萌发、种子耐逆萌发和幼苗建成的分子机理研究进展。
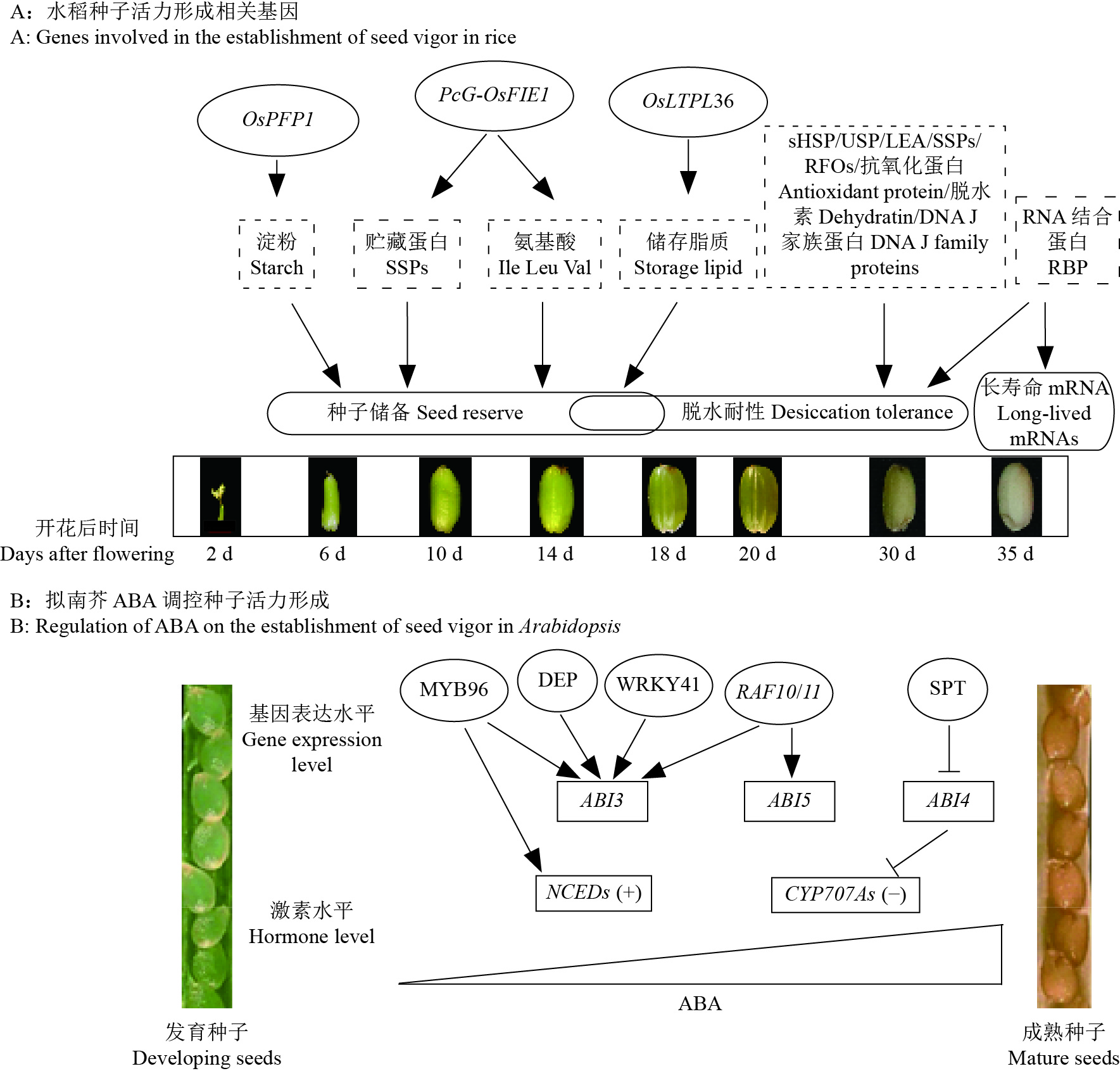
1 种子活力形成的分子机理 1.1 贮藏物质调控机理种子活力形成于发育过程,在此过程中籽粒会积累大量的营养物质,包括碳水化合物、蛋白质和脂类等。在水稻方面,仅有少数几个调控物质积累的基因被报道参与种子活力调控(图1A)。例如,水稻OsPFP1基因突变会显著降低粒质量和淀粉含量,但能显著增加蛋白质和脂质含量,最终会显著减低种子活力[5]。水稻PcG-OsFIE1复合物可调控胚和胚乳发育,影响籽粒贮藏蛋白和氨基酸积累,并参与种子萌发过程[6]。降低水稻脂质转运蛋白编码基因OsLTPL36的表达,会引起籽粒结实率和千粒质量降低,胚乳垩白加重,脂肪酸含量降低,并影响种子萌发[7]。可见,种子发育过程中参与籽粒贮藏物质积累相关的基因是调控种子活力形成的重要因子。
|
图 1 种子发育成熟过程中活力形成的分子机理 Fig. 1 Molecular mechanism on the establishment of seed vigor during seed development and maturation |
种子成熟阶段脱水耐性对种子活力形成具有重要作用。水稻种子在开花后第10—20天开始脱水,第20—40天进入脱水期[8]。已有蛋白质组学研究发现,水稻胚胎发育后期丰富表达蛋白(Late embryogenesis abundant protein,LEA)、小分子热休克蛋白(Small heat shock protein,sHSP)、普遍应激蛋白(Universal stress protein,USP)、抗氧化蛋白、脱水素和DNA J蛋白等,参与种子脱水耐性(图1A)[8-10]。其中,LEA蛋白和可溶性糖协同作用,有助于维持蛋白质和膜结构完整性。在种子成熟过程中,蔗糖和棉子糖(Raffinose,RFO)等非还原性糖逐渐积累,可以防止干燥损伤,在种子脱水耐性形成中发挥着重要作用。目前研究认为,RFOs等物质通过“玻璃化”和“水取代”机制调控种子脱水耐性形成[11]。其中,“玻璃化”机制假说认为,随着失水细胞溶液变得黏稠,这有利于干燥过程中LEA、sHSPs和RFOs等触发“玻璃化”状态,有利于保持细胞稳定性。“水取代”机制假说认为,RFOs羟基可取代细胞内水分子,有利于种子脱水过程中保持天然大分子和膜结构稳定性。此外,在干燥成熟的种子中有些基因mRNA被降解,但也有大量mRNA被储存起来。据报道,在成熟干燥的水稻种子中,超过17000种不同的mRNA被储存起来[12],它们被称为“长寿命mRNA”,即使经历干燥后,它们仍保持活性,可以翻译成蛋白(图1A)。种子吸胀后,储存的mRNA可以被选择性地装载到多聚体中,参与氧化还原、糖酵解和蛋白质合成等过程[13]。但有关长寿命mRNA在种子萌发、休眠、活力调控中的作用仍知之甚少。
1.3 激素调控机理在种子发育成熟过程中,内源激素含量和信号变化,在种子休眠和活力形成中发挥着重要作用(图1B)。在拟南芥中,种子发育成熟过程中脱落酸(Abscisic acid,ABA)含量受合成基因NCEDs和分解基因CYP707A1和CYP707A3的共同调控,影响种子的休眠和萌发[14]。在种子发育过程中,锌指结构域蛋白DESPIERTO(DEP)正向调控ABA信号通路下游重要成分ABI3表达,dep突变体种子休眠特性完全丧失[15]。转录因子WRKY41可以通过直接调控ABI3表达,决定拟南芥种子休眠和萌发[16]。R2R3型转录因子MYB96可以同时调控ABI3及ABA合成基因NCED2和NCED6表达,促进拟南芥种子休眠,抑制种子萌发[17]。MAP3K家族基因RAF10和RAF11通过调节ABI3和ABI5转录水平,调控拟南芥种子休眠[18]。此外,ABI4是ABA信号通路的另一个重要成分,ABI4可以抑制ABA分解相关基因的表达,也是种子休眠调控的重要基因[19];但是ABI4表达受到转录因子SPATULA (SPT)抑制调控[20]。当然,目前所有已知的其他植物激素,如生长素、乙烯、细胞分裂素、赤霉素、油菜素内酯、水杨酸等,也都会参与种子发育成熟过程中的种子活力形成。
2 种子快速萌发的分子机理 2.1 活性氧和激素调控机理作物种子快速、均匀发芽是衡量种子活力的重要指标,对促进植株发育生长,提高产量至关重要。在种子吸胀过程中,过氧化氢(H2O2)通过线粒体呼吸、β−氧化途径、NADPH氧化酶、细胞外过氧化物酶和草酸氧化酶反应等活动产生[21]。研究表明,种子中一些贮藏蛋白和mRNA被氧化有利于促进种子萌发[22-23]。种子萌发过程中产生的ROS通过对贮藏蛋白的氧化修饰有利于贮藏物质的动员[24];且ROS能通过参与胚乳细胞壁的弱化促进种子萌发过程中细胞壁松动,H2O2可以消除ABA对胚乳细胞壁弱化的抑制作用[25]。此外,激素也是调控种子发芽速度的关键因子,目前报道了多个水稻种子活力关键基因通过激素和活性氧调控种子活力(表1)。比如,水稻吲哚乙酸糖基转移酶基因OsIAGLU。通过调控种子萌发过程中生长素(Auxin,IAA)、ABA含量,引起下游ABA信号因子OsABIs表达变化,从而调控水稻种子活力[26]。水稻刺猬互作蛋白类似蛋白OsHIPL1通过ABA代谢和信号途径,影响水孔蛋白OsPIP1;1调节种子萌发过程中水分吸收,提高种子活力[27]。在水稻中,OsRACK1A是最重要的含有WD重复序列的蛋白质家族成员之一,通过控制内源ABA和H2O2水平及其相互作用,正向调控种子萌发[28]。
|
|
表 1 近来报道的控制水稻种子发芽速度相关基因 Table 1 Recently reported genes involving in the speed of seed germination in rice |
近来,多个研究表明在种子萌发过程中氨基酸和能量水平对调控种子活力也具有重要作用。α−异丙基苹果酸合酶OsIPMS是亮氨酸生物合成过程的限速酶,可通过提高水稻种子萌发过程中游离氨基酸的生物合成,促进赤霉素(Gibberellin,GA)合成,从而增强种子萌发过程中糖酵解和三羧酸(Tricarboxylic acid,TCA)循环生化反应,为种子萌发和幼苗生长提供更多能量[29]。水稻Cupin结构域蛋白OsCDP3.10,具有合成相对分子质量为52 000的球蛋白的功能,该基因通过影响种子萌发过程中氨基酸积累,刺激H2O2的产生,从而正向调控种子活力[30]。丙酮酸激酶(Pyruvate kinase,PK)是糖酵解过程中主要的限速酶之一,能够催化磷酸烯醇丙酮酸(Phosphoenolpyruvate,PEP)和二磷酸腺苷(Adenosine diphosphate,ADP)转变为丙酮酸(Pyruvic acid,Pyr)和三磷酸腺苷(Adenosine triphosphate,ATP)。水稻丙酮酸激酶蛋白OsPK5突变后,丙酮酸激酶活性下降,可能通过影响种子萌发过程中糖酵解途径,改变可溶性糖积累,调控植物激素GA/ABA平衡,从而调控种子萌发[31]。水稻OsOMT编码一个2−酮戊二酸/苹果酸转运体蛋白,突变该基因会显著降低种子活力,推测可能是通过调节氨基酸合成、糖酵解和TCA循环过程来影响种子的活力[32]。
2.3 表观遗传调控机理种子萌发过程中,组蛋白修饰介导的表观遗传基因转录调控可能发挥了重要作用,但具体分子机制尚不完全清楚。比如,拟南芥SNL1能够结合组蛋白去乙酰化酶HDA19,调控组蛋白H3K9K18乙酰化水平,影响基因转录;SNL1/SNL2功能缺失影响脱落酸和乙烯相关基因表达,增强了乙烯对脱落酸的拮抗作用,降低了种子休眠[33]。进一步研究发现,SNL1/SNL2功能缺失导致生长素相关基因特别是AUX1的表达升高,增强了生长素在胚根的水平和分布,进而激活下游CYCDs介导的细胞分裂,提高了突变体种子萌发速度[34]。这些研究结果,为解析种子活力调控分子机理提供了重要线索。
3 种子耐逆萌发的分子机理 3.1 种子耐盐萌发分子机理揭示种子耐逆萌发分子机理,对于提升种子活力至关重要,近年来相关研究取得了一定进展(表2)。在种子耐盐萌发分子机理方面,通过图位克隆方法成功克隆了一个高盐胁迫下控制水稻种子快速萌发和幼苗建成的候选基因qSE3,该基因编码一个钾离子转运蛋白OsHAK21,在盐胁迫下qSE3可促进水稻种子萌发过程中K+和Na+的吸收,诱导ABA积累和ABA信号通路基因表达,抑制ROS在种子中的积累,从而提高了种子萌发过程中的耐盐性[35]。水稻OsSAE1可直接与OsABI5启动子结合,通过抑制OsABI5表达,正调控种子耐盐萌发[36]。在拟南芥中,转录因子AtHY5和AtHYH能够与AtRSM1启动子结合,调节其表达,参与盐和ABA胁迫下种子发芽和幼苗生长调控[37]。拟南芥AtABI4可以分别与AtRbohD和AtVTC2结合,调控ROS代谢和细胞膜完整性,参与盐胁迫下种子萌发[38]。拟南芥AtSRT2可调控H2O2囊泡运输相关膜蛋白VAMP714启动子区的组蛋白乙酰化,并抑制VAMP714转录,从而通过改变种子萌发过程中H2O2含量和DNA损伤程度,介导种子萌发期耐盐性[39]。
|
|
表 2 近来报道的控制种子耐逆萌发相关基因 Table 2 Recently reported genes involving in seed germination under stress conditions |
在种子耐冷萌发分子机理研究方面,水稻和拟南芥中已有不少报道(表2)。比如,在种子萌发过程中,水稻种子耐冷萌发基因qLTG3-1在种皮的糊粉层和覆盖胚芽鞘的上胚层表达,能通过调节这些组织的细胞液泡化,从而引起这些组织的松弛而提高种子在低温下的发芽势。在胚胎中强烈表达,有助于组织弱化、降低对胚芽鞘生长的机械阻力,从而促进低温条件下种子发芽速度[40]。水稻OsSAP16正向调控低温下种子萌发,基因表达高低决定了种子耐低温萌发能力[41]。拟南芥AtKP1能够与AtVDAC3特异性相互作用,参与低温条件下种子发芽过程中的呼吸调控作用,Atkp1和Atvdac3突变体发芽种子的耗氧量增加,细胞色素途径和替代氧化酶途径之间的呼吸平衡被破坏,ATP水平降低,进而影响低温下种子活力[42]。此外,AtHSP70-16在质膜和细胞核中与AtVDAC3相互作用,激活AtVDAC3离子通道的开放,从而促进ABA从胚乳流向胚,进而抑制种子发芽[43]。此外,有为数不多的种子耐热萌发机理研究被报道。近来研究发现,拟南芥MADS盒转录因子AGL67能识别SOM启动子区CArG盒,并激活SOM表达,而AGL67招募组蛋白标记阅读器EARLY BOLTING In SHORT DAY(EBS),从而识别SOM染色质上的H3K4me3;在高温条件下,AGL67和EBS在SOM启动子周围高度富集,AGL67-EBS复合物也是组蛋白H4K5乙酰化所必需的,其激活SOM表达,最终抑制种子发芽;该研究揭示了表观遗传学激活SOM表达,以抑制高温胁迫下种子发芽的机制[44]。
3.3 种子耐淹水萌发分子机理在种子耐淹水萌发分子机理研究方面,目前报道主要集中在水稻中(表2)。水稻对淹水胁迫的适应,涉及2种不同的对立机制,即“静止策略”和“逃逸策略”。其中,“逃逸策略”指在淹水条件下,水稻胚芽鞘快速伸长,迅速升至水面,从而将氧气输送到种子中,进行有氧呼吸,对保障直播稻成苗具有重要作用。比如,水稻海藻糖−6−磷酸磷酸酶基因OsTPP7是能量传感器,在淹水条件下,OsTPP7通过增加T6P运转,从而增强淀粉分解以驱动胚和胚芽鞘生长,最终能提升种子萌发过程中的厌氧能力[45]。水稻淹水条件下小分子RNA OsmiR393负调控生长素受体基因OsTIR1和OsAFB2,促进胚芽鞘顶端游离吲哚乙酸(IAA)的积累,从而抑制气孔发育和胚芽鞘伸长[46]。水稻OsCBL10启动子序列变异,决定淹水条件下种子萌发能力,在水稻种子发芽过程中,含耐淹水型OsCBL10启动子品种表现出较低的Ca2+流量和较高的α−淀粉酶活性[47]。淹水条件下,可通过miR167-ARF-GH3途径参与调控游离IAA积累,刺激胚芽鞘伸长,增强水稻种子耐淹水萌发能力[48]。近年来,在杂草稻中鉴定了一个编码14-3-3蛋白基因OsGF14h,其作为ABA信号传导上游信号开关,通过与转录因子OsHOX3和OsVP1互作,维持ABA和GA动态平衡,使厌氧敏感品种在淹水直播条件下出苗率由13.5%提高到60.5%;同时,在人工选择和自然选择中OsGF14h与红米基因Rc协同驯化[49]。水稻OsUGT75A通过糖基化ABA和茉莉酸 (Jasmonic acid,JA),精准调控种子和胚芽鞘中游离态ABA和JA含量,从而通过ABA和JA信号通路介导淹水条件下胚芽鞘伸长[50]。
3.4 种子耐旱萌发分子机理种子耐旱萌发分子机理在番茄和拟南芥中有报道(表2)。在番茄中,转录因子TERF可以激活GA信号通路,负向调控种子发芽过程中对甘露醇处理的敏感性[51]。近来,在拟南芥种子感知水分分子机理研究方面取得重要进展,拟南芥朊病毒样蛋白FLOE1,其在水合作用时相分离,并使植物胚胎能够感知水压力,调控种子发芽最佳时间;研究表明,当休眠种子感应到附近水分时,FLOE1几乎立即聚集在细胞中通过相分离以感应水,以确定条件是否适合种子重新激活并开始生长;FLOE1聚集是暂时和可逆的,它可以作为种子发芽或不发芽的信号,如果确定条件不是最佳的,则停止种子发芽,如果环境有足够的水支持生长,则允许种子继续发芽[52]。
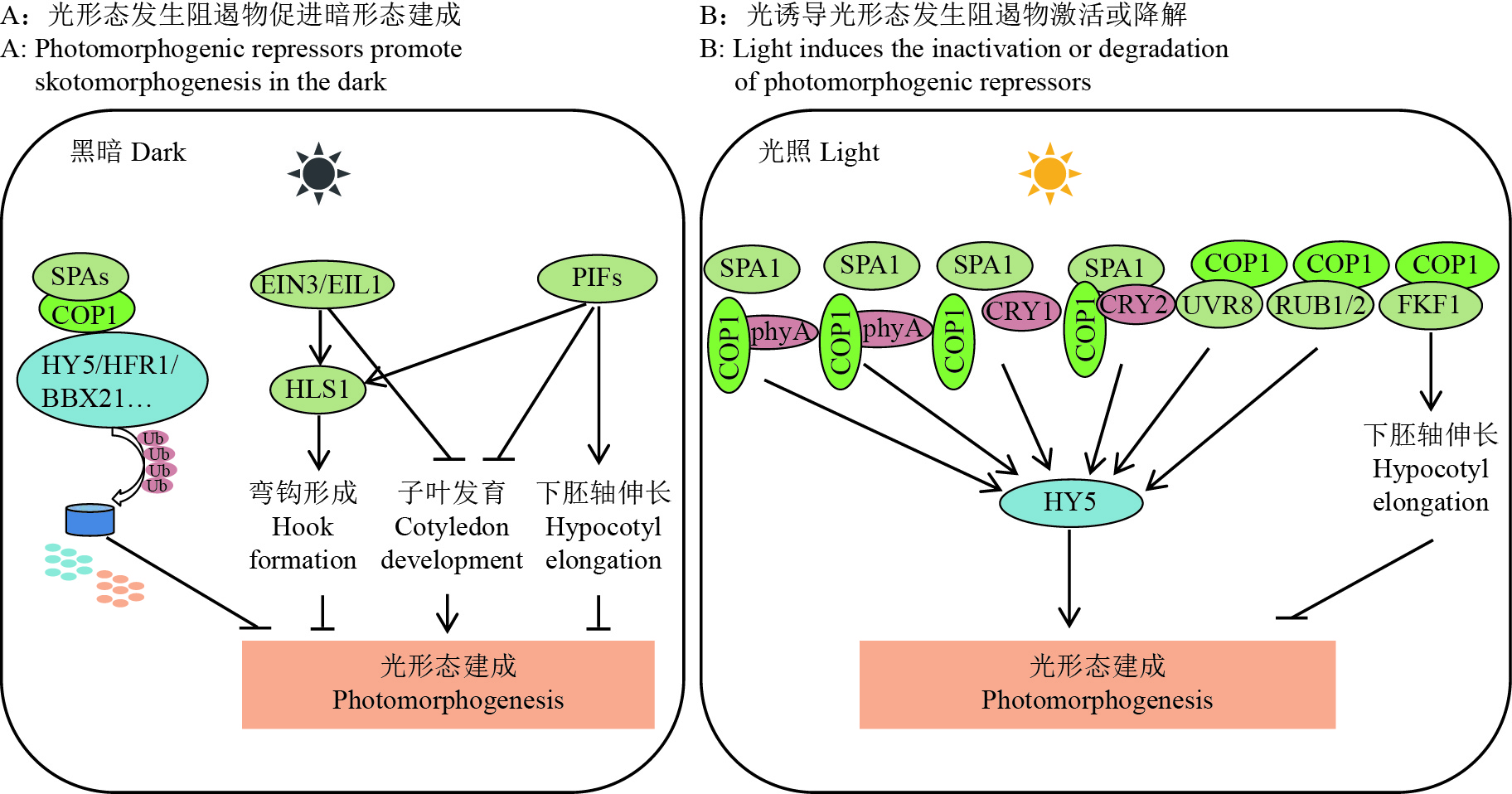
4 种子幼苗建成的分子机理 4.1 光形态建成分子机理幼苗建成是指种子发芽开始,利用自身贮藏物质发育成正常幼苗的过程,当幼苗具备光合能力时,幼苗建成过程就完成了。正常幼苗建成是衡量种子活力水平的重要指标,近年来有关幼苗建成相关分子机制研究取得了长足进展,特别是对拟南芥光形态建成分子机制已有深入研究。在黑暗条件下,拟南芥光感受器处于不活跃状态,光形态抑制因子Constitutively photomorphogenic 1 (COP1)-Suppressor of phytochrome a (SPA)复合物,通过靶向光形态形成促进转录因子,如Elongated hypocotyl 5 (HY5)、Longhypocotyl in far-red 1 (HFR1)、B-box protein 21 (BBX21),进行泛素化和降解,从而促进暗形态发生[53-55]。另一类拟南芥光形态形成阻遏物是bHLH型转录因子Phytochrome-interacting factors (PIFs),通过直接调控下游靶基因表达抑制光形态形成[56]。此外,拟南芥Ethylene-insensitive3(EIN3)和EIN3-like1(EIL1)作为启动乙烯反应转录级联的主要转录因子,已被证明是第3类光形态形成阻遏物,特别是在黑暗条件下弯钩形成和子叶闭合过程[57]。因此,COP1-SPA、PIFs和EIN3/EIL1是在黑暗中维持幼苗黄化表型的主要光形态抑制因子(图2A)。
|
图 2 植物光形态建成分子调控机理模式图 Fig. 2 Model of molecular mechanism of photomorphogenesis in plants |
在光照条件下,活性光感受器对光形态建成阻遏物进行负调控,以促进光形态建成。在红光/远红光反应中,具有生物活性的远红光吸收(Phytochrome far red,Pfr)形式的光敏色素A(Phytochrome A,phyA)和光敏色素B(Phytochrome B,phyB)直接与SPA1结合,破坏COP1和SPAs之间的物理相互作用,导致COP1-SPAs复合物重组,并抑制其泛素化和降解光形态促进转录因子的能力[58-59]。蓝光激活的CRY1与SPA1相互作用,以促进COP1-SPA1复合物的解离,而CRY2与SPA1结合以促进CRY2-COP1的相互作用[60-62]。最新构建的CUL4-DDB1-RUB1/2 E3复合物促进HY5降解,而COP1与RUB1/2相互作用,促进RUP1/2泛素化和降解,以平衡RUP1/2在长时间UV-B诱导的光形态中的积累[63]。UVR8单体与COP1结合,通过阻断COP1和HY5之间的相互作用,以及靶向RUB1/2降解UV-B反应,提高HY5积累。此外,FKF1与COP1的RING结构域相互作用,抑制COP1二聚化,最终使依赖于COP1-SPA复合物的CO降解失活,以调节开花过程,并部分抑制COP1介导的下胚轴伸长[64]。可见,在暴露于光下时,活化的光感受器促进COP1-SPA复合物的解离和COP1二聚化、SPA2降解、COP1从细胞核易位到细胞质,并与COP1底物竞争性结合,降低细胞核中COP1泛素连接酶活性,并允许光形态发生(图2B)。
总之,光形态形成阻遏物,包括COP1、SPAs、CSNs、COP10、DET1、PIFs和EIN3/EIL1,在黑暗中它们具有活性,以促进幼苗的暗形态建成;在光照下,光激发受体与COP1、SPAs、PIFs和EIN3互作,抑制它们的活性或蛋白质积累,以通过增强光形态发生促进因子,如HY5、HYH、LAF1、HFR1、BBX21和GATA2积累,促进植物光形态建成。此外,最近报道了大量的BBX蛋白作为中间体调控光形态发生[65]。
激素和ROS等信号在幼苗建成中也发挥着重要作用。比如,油菜素甾醇(Brassinosteroids,BR)信号参与光介导的幼苗从暗形态建成发育转换到光形态建成;光形态发生抑制子COP1、PIFs和AGB1可增强BR响应,而光形态发生促进因子HY5、BZS1和NF-YCs等可抑制BR信号;BR信号通路基因也参与光信号调控[65]。拟南芥ABA信号终止子(ABA signaling terminator,ABT)与PYR1/PYL/RCAR和PP2C蛋白相互作用,干扰PYR1/PYL4和ABI1/ABI2之间的相互作用,并通过ABA结合PYR1/PYL4阻止对ABI1/AAB2的抑制,从而终止ABA信号传导,这对种子发芽和幼苗建立至关重要[66]。外施H2O2可以在黑暗条件下诱导拟南芥叶片形成,H2O2处理激活光响应基因,这些基因与光合作用、光呼吸和光系统组分相关;植物色素介导的光信号通路参与H2O2促进形态建成过程;组成型光形态发生1和光敏色素相互作用因子3蛋白在H2O2处理条件下下调表达,从而消除了它们在黑暗中对光形态发生的抑制作用[67]。
4.2 激素和环境因子调控机理胚轴伸长是种子萌发后顶土出苗的动力,胚芽鞘为胚体的第一片叶,在种子萌发时,胚芽鞘首先穿出地面,保护胚芽出土时不受损伤。因此,胚轴和胚芽鞘伸长与作物出苗密切相关。除了光形态建成调控胚轴和胚芽鞘生长外,激素和环境因子等对其调控也发挥着重要作用。比如,在水稻中,OsGSK2通过磷酸化调控细胞周期蛋白CYC U2的稳定性,促进细胞分裂[68];OsGY1通过促进JA合成,从而抑制中胚轴和胚芽鞘伸长;而OsEIN2和OsEIL2介导的乙烯信号通路通过抑制GY1及其他JA合成途径基因表达来下调JA含量,促进细胞伸长,进而调控水稻中胚轴和胚芽鞘长度[69]。在拟南芥中,生长素通过MPK3/MPK6调节GRF4蛋白稳定性来增强BZR1核积累,从而促进下胚轴伸长[70];拟南芥合成分子RN1通过TIR1/AFB介导的生长素信号诱导ZAT10、ATL31、WRKY33表达,共同控制下胚轴伸长[71]。
此外,研究发现环境温度可调控拟南芥胚轴生长。拟南芥SMAX1蛋白与phyB相互作用,减轻其对转录因子PIF4的转录活性抑制,促进下胚轴形成;在温暖的条件下,SMAX1蛋白会慢慢失去稳定性,防止下胚轴过度生长[72]。在温暖条件下,拟南芥XBAT31与ELF3互作且泛素化ELF3,并通过26S蛋白酶体促进ELF3降解,促进下胚轴伸长[73]。拟南芥HSP90能增强COP1和ELF3之间的相互作用,减少ELF3对PIF4的功能影响,并在暗形态和热形态发生过程中调节下胚轴伸长[74]。拟南芥热激转录因子HsfA1d是低温下胚轴伸长的正调节因子,在低温条件下,HsfA1d通过促进胞质、质体胞质和质体核糖体蛋白基因的表达,维持低温条件下植物生长的蛋白翻译[75]。类似的,植物激素IAA、ABA、GA、JA、乙烯等在胚芽鞘伸长调控中发挥着重要作用[76-78]。
4.3 能量代谢调控机理植物从种子萌发到幼苗光合系统建立之前是完全异养的,此时需要利用种子中储备的淀粉、糖和蛋白质等维持生命活动的代谢需求。因此,种子发芽后,贮藏物质被分解成可溶性物质,以促进幼苗生长,贮藏物质分解利用效率与幼苗活力相关,幼苗活力是田间幼苗建立和作物产量的关键。三酰甘油(Triacylglycerol,TAG)是双子叶植物种子中储存能量的主要物质,在拟南芥中,转录因子AHL4与磷脂酸互作,调控种子萌发速率、萌发后主根长、种子萌发及苗期建成过程中脂降解的速率,从而影响幼苗建成[79]。在拟南芥中,能量调节因子SnRK1介导拟南芥苗期建成过程中的代谢调控机制,SnRK1的激活会引发大量基因转录和翻译后调节,以抑制胁迫和碳饥饿下的生长并保存能量[80-81];Snrk1α1/α2的缺失导致了贮藏脂质和贮藏蛋白质的降低,从而降低了代谢可用糖和氨基酸的水平,转录因子bZIP63是SnRK1激酶下游靶标,bZIP63被SnRK1磷酸化后,直接靶向并激活cyPPDK启动子,进而在糖异生过程中发挥调控作用[82]。水稻细胞色素b5基因OsCyb5突变体后,成熟种子中α−淀粉酶活性、淀粉和糖动员量显著降低,导致葡萄糖储备不足,萌发过程中物质动员量显著降低,从而影响早期幼苗生长[83]。
5 展望种子活力是由多基因控制的复杂性状,同时受环境影响较大。因此,有必要进一步开发精确鉴定种子活力的方法,将有助于准确揭示其分子机理。在种子发育、贮藏和发芽过程中均会影响种子活力水平,因此需要对种子发育至萌发所有环节进行系统研究。目前,有关种子活力形成、种子快速萌发、种子耐逆萌发、幼苗建成相关分子机理研究,主要集中在拟南芥和水稻等少数模式植物上,有关农作物如玉米、小麦、大豆、油菜等相关研究较少,有待进一步加强。尽管利用各种QTL定位、关联分析、各种组学技术等挖掘了大量种子活力相关的候选基因,但仅有少数基因被开展了研究,总体而言,有关种子活力的分子网络调控仍知之甚少。种子活力相关分子机制的解析,有助于今后培育高活力作物品种,为促进农业增效、农民增收提供帮助。
| [1] |
SUN Q, WANG J H, SUN B Q. Advances on seed vigor physiological and genetic mechanisms[J]. Agricultural Sciences in China, 2007, 6(9): 1060-1066. DOI:10.1016/S1671-2927(07)60147-3 (  0) 0) |
| [2] |
FINCH-SAVAGE W E, BASSEL G W. Seed vigour and crop establishment: Extending performance beyond adaptation[J]. Journal of Experimental Botany, 2016, 67(3): 567-591. DOI:10.1093/jxb/erv490 (  0) 0) |
| [3] |
PELLIZZARO A, NEVEU M, LALANNE D, et al. A role for auxin signaling in the acquisition of longevity during seed maturation[J]. The New Phytologist, 2020, 225(1): 284-296. DOI:10.1111/nph.16150 (  0) 0) |
| [4] |
FAROOQ M, SIDDIQUE K H M, REHMAN H, et al. Rice direct seeding: experiences, challenges and opportunities[J]. Soil & Tillage Research, 2011, 111(2): 87-98. (  0) 0) |
| [5] |
CHEN C, HE B, LIU X, et al. Pyrophosphate-fructose 6-phosphate 1-phosphotransferase (PFP1) regulates starch biosynthesis and seed development via heterotetramer formation in rice (Oryza sativa L. )
[J]. Plant Biotechnology Journal, 2020, 18(1): 83-95. DOI:10.1111/pbi.13173 (  0) 0) |
| [6] |
HUANG X, LU Z, WANG X, et al. Imprinted gene OsFIE1 modulates rice seed development by influencing nutrient metabolism and modifying genome H3K27me3
[J]. The Plant Journal, 2016, 87(3): 305-317. DOI:10.1111/tpj.13202 (  0) 0) |
| [7] |
WANG X, ZHOU W, LU Z, et al. A lipid transfer protein, OsLTPL36, is essential for seed development and seed quality in rice[J]. Plant Science, 2015, 239: 200-208. DOI:10.1016/j.plantsci.2015.07.016 (  0) 0) |
| [8] |
SANO N, MASAKI S, TANABATA T, et al. Proteomic analysis of stress-related proteins in rice seeds during the desiccation phase of grain filling[J]. Plant Biotechnology, 2013, 30(2): 147-156. DOI:10.5511/plantbiotechnology.13.0207a (  0) 0) |
| [9] |
HE D, YANG P. Proteomics of rice seed germination[J]. Frontiers in Plant Science, 2013, 4: 246. (  0) 0) |
| [10] |
WANG W Q, LIU S J, SONG S Q, et al. Proteomics of seed development, desiccation tolerance, germination and vigor[J]. Plant Physiology and Biochemistry, 2015, 86: 1-15. DOI:10.1016/j.plaphy.2014.11.003 (  0) 0) |
| [11] |
SALVI P, VARSHNEY V, MAJEE M. Raffinose family oligosaccharides (RFOs): Role in seed vigor and longevity[J]. Bioscience Reports, 2022, 42(10): BSR20220198. DOI:10.1042/BSR20220198 (  0) 0) |
| [12] |
HOWELL K A, NARSAI R, CARROLL A, et al. Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process[J]. Plant Physiology, 2009, 149(2): 961-980. DOI:10.1104/pp.108.129874 (  0) 0) |
| [13] |
SANO N, RAJJOU L, NORTH H M. Lost in translation: physiological roles of stored mRNAs in seed germination[J]. Plants, 2020, 9(3): 347. DOI:10.3390/plants9030347 (  0) 0) |
| [14] |
PENFIELD S. Seed dormancy and germination[J]. Current Biology, 2017, 27(17): R874-R878. DOI:10.1016/j.cub.2017.05.050 (  0) 0) |
| [15] |
BARRERO J M, MILLAR A A, GRIFFITHS J, et al. Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds
[J]. The Plant Journal, 2010, 61(4): 611-622. DOI:10.1111/j.1365-313X.2009.04088.x (  0) 0) |
| [16] |
DING Z J, YAN J Y, LI G X, et al. WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA
[J]. The Plant Journal, 2014, 79(5): 810-823. DOI:10.1111/tpj.12597 (  0) 0) |
| [17] |
LEE K, LEE H G, YOON S, et al. The Arabidopsis MYB96 transcription factor is a positive regulator of ABSCISIC ACID-INSENSITIVE4 in the control of seed germination
[J]. Plant Physiology, 2015, 168(2): 677-689. DOI:10.1104/pp.15.00162 (  0) 0) |
| [18] |
LEE S, LEE M H, KIM J I, et al. Arabidopsis putative MAP kinase kinase kinases Raf10 and Raf11 are positive regulators of seed dormancy and ABA response
[J]. Plant and Cell Physiology, 2015, 56(1): 84-97. DOI:10.1093/pcp/pcu148 (  0) 0) |
| [19] |
SHU K, ZHANG H, WANG S, et al. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis
[J]. PLoS Genetics, 2013, 9(6): e1003577. DOI:10.1371/journal.pgen.1003577 (  0) 0) |
| [20] |
VAISTIJ F E, GAN Y, PENFIELD S, et al. Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA
[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(26): 10866-10871. (  0) 0) |
| [21] |
WOJTYLA Ł, LECHOWSKA K, KUBALA S, et al. Different modes of hydrogen peroxide action during seed germination[J]. Frontiers in Plant Science, 2016, 7: 66. (  0) 0) |
| [22] |
BARBA-ESPIN G, DIAZ-VIVANCOS P, JOB D, et al. Understanding the role of H2O2 during pea seed germination: a combined proteomic and hormone profiling approach
[J]. Plant, Cell & Environment, 2011, 34(11): 1907-1919. (  0) 0) |
| [23] |
ORACZ K, BOUTEAU H E M, FARRANT J M, et al. ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation[J]. The Plant Journal, 2007, 50(3): 452-465. DOI:10.1111/j.1365-313X.2007.03063.x (  0) 0) |
| [24] |
VERMA G, MISHRA S, SANGWAN N, et al. Reactive oxygen species mediate axis-cotyledon signaling to induce reserve mobilization during germination and seedling establishment in Vigna radiata
[J]. Journal of Plant Physiology, 2015, 184: 79-88. DOI:10.1016/j.jplph.2015.07.001 (  0) 0) |
| [25] |
CHANDRASEKARAN U, ZHAO X, LUO X, et al. Endosperm weakening: The gateway to a seed’ s new life[J]. Plant Physiology and Biochemistry, 2022, 178: 31-39. DOI:10.1016/j.plaphy.2022.02.016 (  0) 0) |
| [26] |
HE Y, ZHAO J, YANG B, et al. Indole-3-acetate beta-glucosyltransferase OsIAGLU regulates seed vigour through mediating crosstalk between auxin and abscisic acid in rice
[J]. Plant Biotechnology Journal, 2020, 18(9): 1933-1945. DOI:10.1111/pbi.13353 (  0) 0) |
| [27] |
HE Y, CHEN S, LIU K, et al. OsHIPL1, a hedgehog-interacting protein-like 1 protein, increases seed vigour in rice[J]. Plant Biotechnology Journal, 2022, 20(7): 1346-1362. DOI:10.1111/pbi.13812 (  0) 0) |
| [28] |
ZHANG D, CHEN L, LI D, et al. OsRACK1 is involved in abscisic acid- and H2O2-mediated signaling to regulate seed germination in rice (Oryza sativa L. )
[J]. PLoS One, 2014, 9(5): e97120. DOI:10.1371/journal.pone.0097120 (  0) 0) |
| [29] |
HE Y, CHENG J, HE Y, et al. Influence of isopropylmalate synthase OsIPMS1 on seed vigour associated with amino acid and energy metabolism in rice
[J]. Plant Biotechnology Journal, 2019, 17(2): 322-337. DOI:10.1111/pbi.12979 (  0) 0) |
| [30] |
PENG L, SUN S, YANG B, et al. Genome-wide association study reveals that the cupin domain protein OsCDP3.10 regulates seed vigour in rice[J]. Plant Biotechnology Journal, 2022, 20(3): 485-498. DOI:10.1111/pbi.13731 (  0) 0) |
| [31] |
YANG B, CHEN M, ZHAN C, et al. Identification of OsPK5 involved in rice glycolytic metabolism and GA/ABA balance for improving seed germination via genome-wide association study
[J]. Journal of Experimental Botany, 2022, 73(11): 3446-3461. DOI:10.1093/jxb/erac071 (  0) 0) |
| [32] |
LI W, YANG B, XU J, et al. A genome-wide association study reveals that the 2-oxoglutarate/malate translocator mediates seed vigor in rice[J]. The Plant Journal, 2021, 108(2): 478-491. DOI:10.1111/tpj.15455 (  0) 0) |
| [33] |
WANG Z, CAO H, SUN Y, et al. Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation
[J]. The Plant Cell, 2013, 25(1): 149-166. DOI:10.1105/tpc.112.108191 (  0) 0) |
| [34] |
WANG Z, CHEN F Y, LI X Y, et al. Arabidopsis seed germination speed is controlled by SNL histone deacetylase-binding factor-mediated regulation of AUX1
[J]. Nature Communications, 2016, 7: 13412. DOI:10.1038/ncomms13412 (  0) 0) |
| [35] |
HE Y, YANG B, HE Y, et al. A quantitative trait locus, qSE3, promotes seed germination and seedling establishment under salinity stress in rice
[J]. The Plant Journal, 2019, 97(6): 1089-1104. DOI:10.1111/tpj.14181 (  0) 0) |
| [36] |
LI Y, ZHOU J, LI Z, et al. Salt and ABA response erf1 improves seed germination and salt tolerance by repressing ABA signaling in rice[J]. Plant Physiology, 2022, 189(2): 1110-1127. DOI:10.1093/plphys/kiac125 (  0) 0) |
| [37] |
YANG B, SONG Z, LI C, et al. RSM1, an Arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity
[J]. PLoS Genetics, 2018, 14(12): e1007839. DOI:10.1371/journal.pgen.1007839 (  0) 0) |
| [38] |
LUO X, DAI Y, ZHENG C, et al. The ABI4-RbohD/VTC2 regulatory module promotes reactive oxygen species (ROS) accumulation to decrease seed germination under salinity stress[J]. The New Phytologist, 2021, 229(2): 950-962. DOI:10.1111/nph.16921 (  0) 0) |
| [39] |
TANG W S, ZHONG L, DING Q Q, et al. Histone deacetylase AtSRT2 regulates salt tolerance during seed germination via repression of vesicle-associated membrane protein 714 (VAMP714) in Arabidopsis
[J]. The New Phytologist, 2022, 234(4): 1278-1293. DOI:10.1111/nph.18060 (  0) 0) |
| [40] |
FUJINO K, SEKIGUCHI H, MATSUDA Y, et al. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice
[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(34): 12623-12628. (  0) 0) |
| [41] |
WANG X, ZOU B, SHAO Q, et al. Natural variation reveals that OsSAP16 controls low-temperature germination in rice[J]. Journal of Experimental Botany, 2018, 69(3): 413-421. DOI:10.1093/jxb/erx413 (  0) 0) |
| [42] |
YANG X Y, CHEN Z W, XU T, et al. Arabidopsis kinesin KP1 specifically interacts with VDAC3, a mitochondrial protein, and regulates respiration during seed germination at low temperature
[J]. The Plant Cell, 2011, 23(3): 1093-1106. DOI:10.1105/tpc.110.082420 (  0) 0) |
| [43] |
ASHRAF M, MAO Q, HONG J, et al. HSP70-16 and VDAC3 jointly inhibit seed germination under cold stress in Arabidopsis
[J]. Plant, Cell & Environment, 2021, 44(11): 3616-3627. (  0) 0) |
| [44] |
LI P, ZHANG Q, HE D, et al. AGAMOUS-LIKE67 cooperates with the histone mark reader EBS to modulate seed germination under high temperature[J]. Plant Physiology, 2020, 184(1): 529-545. DOI:10.1104/pp.20.00056 (  0) 0) |
| [45] |
KRETZSCHMAR T, PELAYO M A, TRIJATMIKO K R, et al. A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice[J]. Nature Plants, 2015, 1: 15124. DOI:10.1038/nplants.2015.124 (  0) 0) |
| [46] |
GUO F, HAN N, XIE Y, et al. The miR393a/target module regulates seed germination and seedling establishment under submergence in rice (Oryza sativa L. )
[J]. Plant, Cell & Environment, 2016, 39(10): 2288-2302. (  0) 0) |
| [47] |
YE N H, WANG F Z, SHI L, et al. Natural variation in the promoter of rice calcineurin B-like protein10 (OsCBL10) affects flooding tolerance during seed germination among rice subspecies
[J]. The Plant Journal, 2018, 94(4): 612-625. DOI:10.1111/tpj.13881 (  0) 0) |
| [48] |
LEE K W, CHEN J J W, WU C S, et al. Auxin plays a role in the adaptation of rice to anaerobic germination and seedling establishment[J]. Plant, Cell & Environment, 2023, 46(4): 1157-1175. (  0) 0) |
| [49] |
SUN J, ZHANG G, CUI Z, et al. Regain flood adaptation in rice through a 14-3-3 protein OsGF14h[J]. Nature Communications, 2022, 13: 5664. DOI:10.1038/s41467-022-33320-x (  0) 0) |
| [50] |
HE Y, SUN S, ZHAO J, et al. UDP-glucosyltransferase OsUGT75A promotes submergence tolerance during rice seed germination[J]. Nature Communications, 2023, 14: 2296. DOI:10.1038/s41467-023-38085-5 (  0) 0) |
| [51] |
LIU H, YUAN L, GUO W, et al. Transcription factor TERF1 promotes seed germination under osmotic conditions by activating gibberellin acid signaling[J]. Plant Science, 2022, 322: 111350. DOI:10.1016/j.plantsci.2022.111350 (  0) 0) |
| [52] |
DORONE Y, BOEYNAEMS S, FLORES E, et al. A prion-like protein regulator of seed germination undergoes hydration-dependent phase separation[J]. Cell, 2021, 184(16): 4284-4298. DOI:10.1016/j.cell.2021.06.009 (  0) 0) |
| [53] |
OSTERLUND M T, HARDTKE C, WEI N, et al. Targeted destabilization of HY5 during light-regulated development of Arabidopsis
[J]. Nature, 2000, 405(6785): 462-466. DOI:10.1038/35013076 (  0) 0) |
| [54] |
XU X, PAIK I, ZHU L, et al. Illuminating progress in phytochrome-mediated light signaling pathways[J]. Trends in Plant Science, 2015, 20(10): 641-650. DOI:10.1016/j.tplants.2015.06.010 (  0) 0) |
| [55] |
XU D, JIANG Y, LI J, et al. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation
[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(27): 7655-7660. (  0) 0) |
| [56] |
LEIVAR P, QUAIL P H. PIFs: Pivotal components in a cellular signaling hub[J]. Trends in Plant Science, 2011, 16(1): 19-28. DOI:10.1016/j.tplants.2010.08.003 (  0) 0) |
| [57] |
SHI H, LYU M, LUO Y, et al. Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(25): 6482-6487. (  0) 0) |
| [58] |
LU X D, ZHOU C M, XU P B, et al. Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis
[J]. Molecular Plant, 2015, 8(3): 467-478. DOI:10.1016/j.molp.2014.11.025 (  0) 0) |
| [59] |
SHEERIN D J, MENON C, ZUR OVEN-KROCKHAUS S, et al. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex
[J]. The Plant Cell, 2015, 27(1): 189-201. DOI:10.1105/tpc.114.134775 (  0) 0) |
| [60] |
ZUO Z, LIU H, LIU B, et al. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis
[J]. Current Biology, 2011, 21(10): 841-847. DOI:10.1016/j.cub.2011.03.048 (  0) 0) |
| [61] |
LIU B, ZUO Z, LIU H, et al. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light
[J]. Genes & Development, 2011, 25(10): 1029-1034. (  0) 0) |
| [62] |
LIAN H L, HE S B, ZHANG Y C, et al. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism[J]. Genes & Development, 2011, 25(10): 1023-1028. (  0) 0) |
| [63] |
REN H, HAN J, YANG P, et al. Two E3 ligases antagonistically regulate the UV-B response in Arabidopsis
[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(10): 4722-4731. (  0) 0) |
| [64] |
LEE B D, CHA J Y, KIM M R, et al. Photoperiod sensing system for timing of flowering in plants[J]. BMB Reports, 2018, 51(4): 163-164. DOI:10.5483/BMBRep.2018.51.4.052 (  0) 0) |
| [65] |
LIN F, CAO J, YUAN J, et al. Integration of light and brassinosteroid signaling during seedling establishment[J]. International Journal of Molecular Sciences, 2021, 22(23): 12971. DOI:10.3390/ijms222312971 (  0) 0) |
| [66] |
WANG Z, REN Z, CHENG C, et al. Counteraction of ABA-mediated inhibition of seed germination and seedling establishment by ABA signaling terminator in Arabidopsis
[J]. Molecular Plant, 2020, 13(9): 1284-1297. DOI:10.1016/j.molp.2020.06.011 (  0) 0) |
| [67] |
CHENG H, LIANG Q, CHEN X, et al. Hydrogen peroxide facilitates Arabidopsis seedling establishment by interacting with light signalling pathway in the dark
[J]. Plant, Cell & Environment, 2019, 42(4): 1302-1317. (  0) 0) |
| [68] |
SUN S, WANG T, WANG L, et al. Natural selection of a GSK3 determines rice mesocotyl domestication by coordinating strigolactone and brassinosteroid signaling[J]. Nature Communications, 2018, 9(1): 2523. DOI:10.1038/s41467-018-04952-9 (  0) 0) |
| [69] |
XIONG Q, MA B, LU X, et al. Ethylene-inhibited jasmonic acid biosynthesis promotes mesocotyl/coleoptile elongation of etiolated rice seedlings[J]. The Plant Cell, 2017, 29(5): 1053-1072. DOI:10.1105/tpc.16.00981 (  0) 0) |
| [70] |
YU Z, MA J, ZHANG M, et al. Auxin promotes hypocotyl elongation by enhancing BZR1 nuclear accumulation in Arabidopsis
[J]. Science Advances, 2023, 9(1): eade2493. DOI:10.1126/sciadv.ade2493 (  0) 0) |
| [71] |
RIGAL A, DOYLE S M, RITTER A, et al. A network of stress-related genes regulates hypocotyl elongation downstream of selective auxin perception[J]. Plant Physiology, 2021, 187(1): 430-445. DOI:10.1093/plphys/kiab269 (  0) 0) |
| [72] |
PARK Y J, KIM J Y, PARK C M. SMAX1 potentiates phytochrome B-mediated hypocotyl thermomorphogenesis[J]. The Plant Cell, 2022, 34(7): 2671-2687. DOI:10.1093/plcell/koac124 (  0) 0) |
| [73] |
ZHANG L L, SHAO Y J, DING L, et al. XBAT31 regulates thermoresponsive hypocotyl growth through mediating degradation of the thermosensor ELF3 in Arabidopsis
[J]. Science Advances, 2021, 7(19): eabf4427. DOI:10.1126/sciadv.abf4427 (  0) 0) |
| [74] |
ZENG Y, WANG J, HUANG S, et al. HSP90s are required for hypocotyl elongation during skotomorphogenesis and thermomorphogenesis via the COP1-ELF3-PIF4 pathway in Arabidopsis
[J]. The New Phytologist, 2023, 239(4): 1253-1265. DOI:10.1111/nph.18776 (  0) 0) |
| [75] |
LIU H, ZHANG Y, LU S, et al. HsfA1d promotes hypocotyl elongation under chilling via enhancing expression of ribosomal protein genes in Arabidopsis
[J]. The New Phytologist, 2021, 231(2): 646-660. DOI:10.1111/nph.17413 (  0) 0) |
| [76] |
NGHI K N, TONDELLI A, VALE G, et al. Dissection of coleoptile elongation in japonica rice under submergence through integrated genome-wide association mapping and transcriptional analyses
[J]. Plant, Cell & Environment, 2019, 42(6): 1832-1846. (  0) 0) |
| [77] |
NGHI K N, TAGLIANI A, MARIOTTI L, et al. Auxin is required for the long coleoptile trait in japonica rice under submergence[J]. New Phytologist, 2021, 229(1): 85-93. DOI:10.1111/nph.16781 (  0) 0) |
| [78] |
YIN C C, HUANG Y H, ZHANG X, et al. Ethylene-mediated regulation of coleoptile elongation in rice seedlings[J]. Plant, Cell & Environment, 2023, 46(4): 1060-1074. (  0) 0) |
| [79] |
CAI G, KIM S C, LI J, et al. Transcriptional regulation of lipid catabolism during seedling establishment[J]. Molecular Plant, 2020, 13(7): 984-1000. DOI:10.1016/j.molp.2020.04.007 (  0) 0) |
| [80] |
CREPIN N, ROLLAND F. SnRK1 activation, signaling, and networking for energy homeostasis[J]. Current Opinion in Plant Biology, 2019, 51: 29-36. DOI:10.1016/j.pbi.2019.03.006 (  0) 0) |
| [81] |
BAENA-GONZALEZ E, SHEEN J. Convergent energy and stress signaling[J]. Trends in Plant Science, 2008, 13(9): 474-482. DOI:10.1016/j.tplants.2008.06.006 (  0) 0) |
| [82] |
HENNINGER M, PEDROTTI L, KRISCHKE M, et al. The evolutionarily conserved kinase SnRK1 orchestrates resource mobilization during Arabidopsis seedling establishment
[J]. The Plant Cell, 2022, 34(1): 616-632. DOI:10.1093/plcell/koab270 (  0) 0) |
| [83] |
HUANG Z, YING J, PENG L, et al. A genome-wide association study reveals that the cytochrome b5 involved in seed reserve mobilization during seed germination in rice
[J]. Theoretical and Applied Genetics, 2021, 134(12): 4067-4076. DOI:10.1007/s00122-021-03948-2 (  0) 0) |
 2023, Vol. 44
2023, Vol. 44


